SB202190 Predicts BRAF-Activating Mutations in Primary Colorectal Cancer Organoids via Erk1-2 Modulation
- PMID: 36831331
- PMCID: PMC9954675
- DOI: 10.3390/cells12040664
SB202190 Predicts BRAF-Activating Mutations in Primary Colorectal Cancer Organoids via Erk1-2 Modulation
Abstract
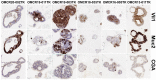
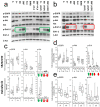
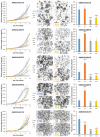
The p38 inhibitor SB202190 is a necessary component of the medium used for normal colorectal mucosa cultures. Sato et al. suggested that the primary activity of SB202190 may be EGFR signaling stabilization, causing an increased phosphorylation of Erk1-2 sustaining organoid proliferation. However, the growth of some colorectal cancer (CRC)-derived organoid cultures is inhibited by this molecule via an unknown mechanism. We biochemically investigated SB202190 activity on a collection of 25 primary human CRC organoids, evaluating EGFR, Akt and Erk1-2 activation using Western blot. We found that Erk1-2 phosphorylation was induced by SB202190 in 20 organoid cultures and inhibited in 5 organoid cultures. A next-generation sequencing (NGS) analysis revealed that the inhibition of p-Erk1-2 signaling corresponded to the cultures with BRAF mutations (with four different hits, one being undescribed), while p-Erk1-2 induction was apparently unrelated to other mutations involving the EGFR pathway (Her2, KRAS and NRAS). We found that SB202190 mirrored the biochemical activity of the BRAF inhibitor Dabrafenib, known to induce the paradoxical activation of p-Erk1-2 signaling in BRAF wild-type cells. SB202190 was a more effective inhibitor of BRAF-mutated organoid growth in the long term than the specific BRAF inhibitors Dabrafenib and PLX8394. Overall, SB202190 can predict BRAF-activating mutations in patient-derived organoids, as well as allowing for the identification of new BRAF variants, preceding and enforcing NGS data.
Keywords: BRAF; Erk1-2; SB202190; colorectal cancer; organoid.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


Similar articles
-
Recommendations from the EGAPP Working Group: can testing of tumor tissue for mutations in EGFR pathway downstream effector genes in patients with metastatic colorectal cancer improve health outcomes by guiding decisions regarding anti-EGFR therapy?Genet Med. 2013 Jul;15(7):517-27. doi: 10.1038/gim.2012.184. Epub 2013 Feb 21. Genet Med. 2013. PMID: 23429431
-
Tumor suppressor miR-193a-3p enhances efficacy of BRAF/MEK inhibitors in BRAF-mutated colorectal cancer.Cancer Sci. 2021 Sep;112(9):3856-3870. doi: 10.1111/cas.15075. Epub 2021 Jul 29. Cancer Sci. 2021. PMID: 34288281 Free PMC article.
-
An amino-terminal BRAF deletion accounting for acquired resistance to RAF/EGFR inhibition in colorectal cancer.Cold Spring Harb Mol Case Stud. 2020 Aug 25;6(4):a005140. doi: 10.1101/mcs.a005140. Print 2020 Aug. Cold Spring Harb Mol Case Stud. 2020. PMID: 32669268 Free PMC article.
-
BGB-283, a Novel RAF Kinase and EGFR Inhibitor, Displays Potent Antitumor Activity in BRAF-Mutated Colorectal Cancers.Mol Cancer Ther. 2015 Oct;14(10):2187-97. doi: 10.1158/1535-7163.MCT-15-0262. Epub 2015 Jul 24. Mol Cancer Ther. 2015. PMID: 26208524
-
Targeting oncogenic Raf protein-serine/threonine kinases in human cancers.Pharmacol Res. 2018 Sep;135:239-258. doi: 10.1016/j.phrs.2018.08.013. Epub 2018 Aug 15. Pharmacol Res. 2018. PMID: 30118796 Review.
Cited by
-
Epidermal Growth Factor Receptor Targeting in Colorectal Carcinoma: Antibodies and Patient-Derived Organoids as a Smart Model to Study Therapy Resistance.Int J Mol Sci. 2024 Jun 28;25(13):7131. doi: 10.3390/ijms25137131. Int J Mol Sci. 2024. PMID: 39000238 Free PMC article. Review.
-
Cyclic fasting bolsters cholesterol biosynthesis inhibitors' anticancer activity.Nat Commun. 2023 Oct 31;14(1):6951. doi: 10.1038/s41467-023-42652-1. Nat Commun. 2023. PMID: 37907500 Free PMC article.
-
Modelling esophageal adenocarcinoma and Barrett's esophagus with patient-derived organoids.Front Mol Biosci. 2024 Apr 24;11:1382070. doi: 10.3389/fmolb.2024.1382070. eCollection 2024. Front Mol Biosci. 2024. PMID: 38721276 Free PMC article. Review.
-
Ex Vivo Intestinal Organoid Models: Current State-of-the-Art and Challenges in Disease Modelling and Therapeutic Testing for Colorectal Cancer.Cancers (Basel). 2024 Oct 30;16(21):3664. doi: 10.3390/cancers16213664. Cancers (Basel). 2024. PMID: 39518102 Free PMC article. Review.
-
High-throughput solutions in tumor organoids: from culture to drug screening.Stem Cells. 2025 Jan 17;43(1):sxae070. doi: 10.1093/stmcls/sxae070. Stem Cells. 2025. PMID: 39460616 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

