Multi-Omics Integration Analysis of TK1 in Glioma: A Potential Biomarker for Predictive, Preventive, and Personalized Medical Approaches
- PMID: 36831773
- PMCID: PMC9954725
- DOI: 10.3390/brainsci13020230
Multi-Omics Integration Analysis of TK1 in Glioma: A Potential Biomarker for Predictive, Preventive, and Personalized Medical Approaches
Abstract
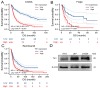
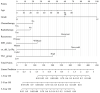
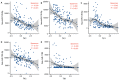
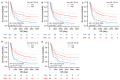
Multi-omics expression datasets obtained from multiple public databases were used to elucidate the biological function of TK1 and its effects on clinical outcomes. The Kaplan-Meier curve, a predictive nomogram mode, and the time-dependent receiver operating characteristic (ROC) curve were established to assess the role of TK1 expression in glioma prognosis. TK1 was overexpressed in glioma compared with normal samples, and patients with elevated expression of TK1 had poor overall survival. The ROC curves indicated a high diagnostic value of TK1 expression in patients of glioma; the areas under the ROC curve (AUC) were 0.682, 0.735, and 0.758 for 1 year, 3 years, and 5 years of glioma survival, respectively. For a model based on TK1 expression and other clinical characteristics, the values of AUC were 0.864, 0.896, and 0.898 for 1 year, 3 years, and 5 years, respectively. Additionally, the calibration curve indicated that the predicted and observed areas at 1 year, 3 years, and 5 years of survival were in excellent agreement. Three types of TK1 alterations-missense mutations, splice mutations, and amplifications-were identified in 25 of 2706 glioma samples. The TK1-altered group had better overall survival than the unaltered group. Single-cell function analysis showed that TK1 was positively associated with proliferation, the cell cycle, DNA repair, DNA damage, and epithelial-mesenchymal transition in glioma. Immunoinfiltration analysis indicated that TK1 expression might play different roles in low-grade glioma and glioblastoma multiforme tumor microenvironments, but TK1 expression was positively associated with activated CD4 and Th2, regardless of tumor grade. In summary, our findings identified TK1 as a novel marker for predicting clinical outcomes and a potential target for glioma.
Keywords: TK1; bioinformatics; glioma; medical informatics; multi-omics integration analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Cabel L., Rosenblum D., Lerebours F., Brain E., Loirat D., Bergqvist M., Cottu P., Donnadieu A., Bethune A., Kiavue N., et al. Plasma thymidine kinase 1 activity and outcome of ER+ HER2- metastatic breast cancer patients treated with palbociclib and endocrine therapy. Breast Cancer Res. 2020;22:98. doi: 10.1186/s13058-020-01334-2. - DOI - PMC - PubMed
-
- Liu C., Wang J., Zhao L., He H., Zhao P., Peng Z., Liu F., Chen J., Wu W., Wang G., et al. Knockdown of Thymidine Kinase 1 Suppresses Cell Proliferation; Invasion; Migration; and Epithelial-Mesenchymal Transition in Thyroid Carcinoma Cells. Front. Oncol. 2020;9:1475. doi: 10.3389/fonc.2019.01475. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

