Performance of T-Track® TB, a Novel Dual Marker RT-qPCR-Based Whole-Blood Test for Improved Detection of Active Tuberculosis
- PMID: 36832246
- PMCID: PMC9955725
- DOI: 10.3390/diagnostics13040758
Performance of T-Track® TB, a Novel Dual Marker RT-qPCR-Based Whole-Blood Test for Improved Detection of Active Tuberculosis
Abstract
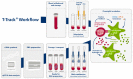
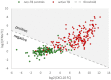
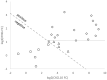
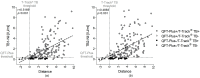
Tuberculosis (TB) is one of the leading causes of death by an infectious disease. It remains a major health burden worldwide, in part due to misdiagnosis. Therefore, improved diagnostic tests allowing the faster and more reliable diagnosis of patients with active TB are urgently needed. This prospective study examined the performance of the new molecular whole-blood test T-Track® TB, which relies on the combined evaluation of IFNG and CXCL10 mRNA levels, and compared it to that of the QuantiFERON®-TB Gold Plus (QFT-Plus) enzyme-linked immunosorbent assay (ELISA). Diagnostic accuracy and agreement analyses were conducted on the whole blood of 181 active TB patients and 163 non-TB controls. T-Track® TB presented sensitivity of 94.9% and specificity of 93.8% for the detection of active TB vs. non-TB controls. In comparison, the QFT-Plus ELISA showed sensitivity of 84.3%. The sensitivity of T-Track® TB was significantly higher (p < 0.001) than that of QFT-Plus. The overall agreement of T-Track® TB with QFT-Plus to diagnose active TB was 87.9%. Out of 21 samples with discordant results, 19 were correctly classified by T-Track® TB while misclassified by QFT-Plus (T-Track® TB-positive/QFT-Plus-negative), and two samples were misclassified by T-Track® TB while correctly classified by QFT-Plus (T-Track® TB-negative/QFT-Plus-positive). Our results demonstrate the excellent performance of the T-Track® TB molecular assay and its suitability to accurately detect TB infection and discriminate active TB patients from non-infected controls.
Keywords: CXCL10; IFNG; QuantiFERON®-TB Gold Plus; RT-qPCR; T-Track® TB; TB; active TB; infection detection; mRNA; tuberculosis.
Conflict of interest statement
J.P.M., S.M., F.H., L.D. and S.B. are employees of Mikrogen. J.P.M., S.M., F.H., A.A.-N., L.D. and S.B. were employees of Lophius Biosciences (meanwhile acquired by Mikrogen). L.D. is the co-founder and Chief Scientific Officer of Lophius Biosciences. R.W. is Chairman of the Board of Lophius Biosciences. R.W., L.D. and S.B. are shareholders of Lophius Biosciences. The participating clinical and measurement centers received research funding from Lophius Biosciences for this study. Lophius Biosciences and then Mikrogen played a role in the design of the study, in the collection, analysis and interpretation of data, in the decision to publish the results, and in the writing of the manuscript.
Figures
References
-
- World Health Organization (WHO) Tuberculosis (TB) [(accessed on 3 November 2022)]. Available online: https://www.who.int/news-room/fact-sheets/detail/tuberculosis.
-
- TB Alliance Global Pandemic. [(accessed on 3 November 2022)]. Available online: https://www.tballiance.org/why-new-tb-drugs/global-pandemic.
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

