Rapid Quantitative Method Development for Beef and Pork Lymph Nodes Using BAX® System Real Time Salmonella Assay
- PMID: 36832897
- PMCID: PMC9956926
- DOI: 10.3390/foods12040822
Rapid Quantitative Method Development for Beef and Pork Lymph Nodes Using BAX® System Real Time Salmonella Assay
Abstract
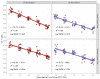
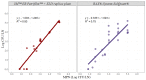
The goal of this study was to develop a rapid RT-PCR enumeration method for Salmonella in pork and beef lymph nodes (LNs) utilizing BAX®-System-SalQuant® as well as to assess the performance of the methodology in comparison with existing ones. For study one: PCR curve development, pork, and beef LNs (n = 64) were trimmed, sterilized, pulverized, spiked with 0.00 to 5.00 Log CFU/LN using Salmonella Typhimurium, and then homogenized with BAX-MP media. Samples were incubated at 42 °C and tested at several time points using the BAX®-System-RT-PCR Assay for Salmonella. Cycle-Threshold values from the BAX®-System, for each Salmonella concentration were recorded and utilized for statistical analysis. For study two: Method comparison; additional pork and beef LNs (n = 52) were spiked and enumerated by (1) 3M™EB-Petrifilm™ + XLD-replica plate, (2) BAX®-System-SalQuant®, and (3) MPN. Linear-fit equations for LNs were estimated with recovery times of 6 h and a limit of quantification (LOQ) of 10 CFU/LN. Slopes and intercepts for LNs using BAX®-System-SalQuant® when compared with MPN were not significantly different (p < 0.05), while the same parameters for 3M™EB-Petrifilm™ + XLD-replica plate were significantly different (p > 0.05). The results support the capability of BAX®-System-SalQuant® to enumerate Salmonella in pork and beef LNs. This development adds support to the use of PCR-based quantification methodologies for pathogen loads in meat products.
Keywords: Salmonella quantification; beef lymph nodes; pathogen control; pork lymph nodes.
Conflict of interest statement
The authors declare no conflict of interest. The company had no role in the collection, analysis, interpretation of data conclusions nor in the writing of this article. The company contributed with supplies to develop the methodology.
Figures


References
-
- Centers for Disease Control and Prevention Surveillance for Foodborne Disease Outbreaks—United States, 2017: Annual Report; Atlanta, 2019. [(accessed on 10 October 2022)]; Available online: https://www.cdc.gov/fdoss/annual-reports/index.html.
-
- Interagency Food Safety Analytics Collaboration Foodborne Illness Source Attribution Estimates for 2018 for Salmonella, Escherichia Coli O157, Listeria Monocytogenes, and Campylobacter Using Multi-Year Outbreak Surveillance Data, United States. [(accessed on 10 October 2022)];2020 Available online: https://www.cdc.gov/foodsafety/ifsac/annual-reports.html.
-
- Food Safety and Inspection Service Changes to the Salmonella Verification Testing Program: Proposed Performance Standards for Salmonella in Raw Comminuted Pork and Intact or Non-Intact Pork Cuts and Related Agency Verification Procedures. [(accessed on 8 October 2022)];2022 Available online: https://www.fsis.usda.gov/policy/federal-register-rulemaking/federal-reg....
-
- Centers for Disease Control and Prevention Salmonella and Food. [(accessed on 5 October 2022)];2022 Available online: https://www.cdc.gov/foodsafety/communication/salmonella-food.html.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

