Haploinsufficiency as a Foreground Pathomechanism of Poirer-Bienvenu Syndrome and Novel Insights Underlying the Phenotypic Continuum of CSNK2B-Associated Disorders
- PMID: 36833176
- PMCID: PMC9957394
- DOI: 10.3390/genes14020250
Haploinsufficiency as a Foreground Pathomechanism of Poirer-Bienvenu Syndrome and Novel Insights Underlying the Phenotypic Continuum of CSNK2B-Associated Disorders
Abstract
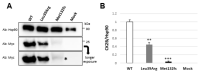
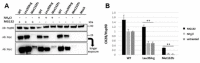
CSNK2B encodes for the regulatory subunit of the casein kinase II, a serine/threonine kinase that is highly expressed in the brain and implicated in development, neuritogenesis, synaptic transmission and plasticity. De novo variants in this gene have been identified as the cause of the Poirier-Bienvenu Neurodevelopmental Syndrome (POBINDS) characterized by seizures and variably impaired intellectual development. More than sixty mutations have been described so far. However, data clarifying their functional impact and the possible pathomechanism are still scarce. Recently, a subset of CSNK2B missense variants affecting the Asp32 in the KEN box-like domain were proposed as the cause of a new intellectual disability-craniodigital syndrome (IDCS). In this study, we combined predictive functional and structural analysis and in vitro experiments to investigate the effect of two CSNK2B mutations, p.Leu39Arg and p.Met132LeufsTer110, identified by WES in two children with POBINDS. Our data prove that loss of the CK2beta protein, due to the instability of mutant CSNK2B mRNA and protein, resulting in a reduced amount of CK2 complex and affecting its kinase activity, may underlie the POBINDS phenotype. In addition, the deep reverse phenotyping of the patient carrying p.Leu39Arg, with an analysis of the available literature for individuals with either POBINDS or IDCS and a mutation in the KEN box-like motif, might suggest the existence of a continuous spectrum of CSNK2B-associated phenotypes rather than a sharp distinction between them.
Keywords: CK2beta haploinsufficiency; IDCS; KEN like-box; POBINDS; WES; mRNA and protein instability; phenotypic continuum.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

