Stage-Specific Transcriptomes of the Mussel Mytilus coruscus Reveals the Developmental Program for the Planktonic to Benthic Transition
- PMID: 36833215
- PMCID: PMC9957406
- DOI: 10.3390/genes14020287
Stage-Specific Transcriptomes of the Mussel Mytilus coruscus Reveals the Developmental Program for the Planktonic to Benthic Transition
Abstract
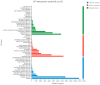
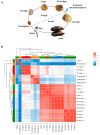
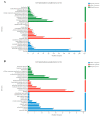
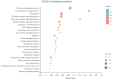
Many marine invertebrate larvae undergo complex morphological and physiological changes during the planktonic-benthic transition (a.k.a. metamorphosis). In this study, transcriptome analysis of different developmental stages was used to uncover the molecular mechanisms underpinning larval settlement and metamorphosis of the mussel, Mytilus coruscus. Analysis of highly upregulated differentially expressed genes (DEGs) at the pediveliger stage revealed enrichment of immune-related genes. The results may indicate that larvae co-opt molecules of the immune system to sense and respond to external chemical cues and neuroendocrine signaling pathways forecast and trigger the response. The upregulation of adhesive protein genes linked to byssal thread secretion indicates the anchoring capacity required for larval settlement arises prior to metamorphosis. The results of gene expression support a role for the immune and neuroendocrine systems in mussel metamorphosis and provide the basis for future studies to disentangle gene networks and the biology of this important lifecycle transformation.
Keywords: Mytilus coruscus; hard-shelled mussel; larval settlement and metamorphosis; pediveliger larvae; transcriptome.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


Similar articles
-
Thyroid hormone receptor: A new player in epinephrine-induced larval metamorphosis of the hard-shelled mussel.Gen Comp Endocrinol. 2020 Feb 1;287:113347. doi: 10.1016/j.ygcen.2019.113347. Epub 2019 Nov 30. Gen Comp Endocrinol. 2020. PMID: 31794730
-
Hedgehog signaling is required for larval muscle development and larval metamorphosis of the mussel Mytilus coruscus.Dev Biol. 2024 Aug;512:57-69. doi: 10.1016/j.ydbio.2024.05.007. Epub 2024 May 13. Dev Biol. 2024. PMID: 38750688
-
An ɑ2-adrenergic receptor is involved in larval metamorphosis in the mussel, Mytilus coruscus.Biofouling. 2019 Oct;35(9):986-996. doi: 10.1080/08927014.2019.1685661. Epub 2019 Nov 14. Biofouling. 2019. PMID: 31724449
-
Expansion and diversity of caspases in Mytilus coruscus contribute to larval metamorphosis and environmental adaptation.BMC Genomics. 2024 Mar 27;25(1):314. doi: 10.1186/s12864-024-10238-w. BMC Genomics. 2024. PMID: 38532358 Free PMC article. Review.
-
The initiation of metamorphosis as an ancient polyphenic trait and its role in metazoan life-cycle evolution.Philos Trans R Soc Lond B Biol Sci. 2010 Feb 27;365(1540):641-51. doi: 10.1098/rstb.2009.0248. Philos Trans R Soc Lond B Biol Sci. 2010. PMID: 20083639 Free PMC article. Review.
Cited by
-
Gonadal Lipid Storage in Mytilus coruscus: A Comprehensive Gene Network and Key Gene Discovery.Mar Biotechnol (NY). 2025 Jun 19;27(4):99. doi: 10.1007/s10126-025-10475-4. Mar Biotechnol (NY). 2025. PMID: 40536640
-
Identification of the principal neuropeptide MIP and its action pathway in larval settlement of the echiuran worm Urechis unicinctus.BMC Genomics. 2024 Apr 3;25(1):337. doi: 10.1186/s12864-024-10228-y. BMC Genomics. 2024. PMID: 38641568 Free PMC article.
-
ChIP-seq identifies McSLC35E2 as a novel target gene of McNrf2 in Mytilus coruscus, highlighting its role in the regulation of oxidative stress response in marine mollusks.Front Physiol. 2023 Oct 6;14:1282900. doi: 10.3389/fphys.2023.1282900. eCollection 2023. Front Physiol. 2023. PMID: 37869713 Free PMC article.
-
Balancing selection and candidate loci for survival and growth during larval development in the Mediterranean mussel, Mytilus galloprovincialis.G3 (Bethesda). 2023 Jul 5;13(7):jkad103. doi: 10.1093/g3journal/jkad103. G3 (Bethesda). 2023. PMID: 37178422 Free PMC article.
References
-
- China Fishery Statistical Yearbook. China Agriculture Press; Beijing, China: 2022. Bureau of Fisheries of the Ministry of Agriculture of the People’s Republic of China (BFMOA) p. 43. (In Chinese)
-
- Li Y.F., Cheng Y.L., Chen K., Cheng Z.Y., Zhu X., Cardoso J.C.R., Liang X., Zhu Y.T., Power D.M., Yang J.L. Thyroid hormone receptor: A new player in epinephrine-induced larval metamorphosis of the hard-shelled mussel. Gen. Comp. Endocrinol. 2020;287:113347–113355. doi: 10.1016/j.ygcen.2019.113347. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

