RAD51 and RAD51B Play Diverse Roles in the Repair of DNA Double Strand Breaks in Physcomitrium patens
- PMID: 36833232
- PMCID: PMC9956106
- DOI: 10.3390/genes14020305
RAD51 and RAD51B Play Diverse Roles in the Repair of DNA Double Strand Breaks in Physcomitrium patens
Abstract
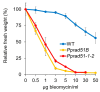
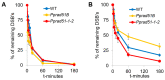
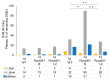
RAD51 is involved in finding and invading homologous DNA sequences for accurate homologous recombination (HR). Its paralogs have evolved to regulate and promote RAD51 functions. The efficient gene targeting and high HR rates are unique in plants only in the moss Physcomitrium patens (P. patens). In addition to two functionally equivalent RAD51 genes (RAD1-1 and RAD51-2), other RAD51 paralogues were also identified in P. patens. For elucidation of RAD51's involvement during DSB repair, two knockout lines were constructed, one mutated in both RAD51 genes (Pprad51-1-2) and the second with mutated RAD51B gene (Pprad51B). Both lines are equally hypersensitive to bleomycin, in contrast to their very different DSB repair efficiency. Whereas DSB repair in Pprad51-1-2 is even faster than in WT, in Pprad51B, it is slow, particularly during the second phase of repair kinetic. We interpret these results as PpRAD51-1 and -2 being true functional homologs of ancestral RAD51 involved in the homology search during HR. Absence of RAD51 redirects DSB repair to the fast NHEJ pathway and leads to a reduced 5S and 18S rDNA copy number. The exact role of the RAD51B paralog remains unclear, though it is important in damage recognition and orchestrating HR response.
Keywords: DNA double-strand break (DSB); Physcomitrella; bleomycin; comet assay; evolutionary divergence; homologous recombination (HR); non-homologous end-joining (NHEJ); rDNA; repair kinetic.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Aboussekhra A., Chanet R., Adjiri A., Fabre F. Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol. Cell. Biol. 1992;12:3224–3234. doi: 10.1128/mcb.12.7.3224-3234.1992. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

