Characterization and Expression of Holothurian Wnt Signaling Genes during Adult Intestinal Organogenesis
- PMID: 36833237
- PMCID: PMC9957329
- DOI: 10.3390/genes14020309
Characterization and Expression of Holothurian Wnt Signaling Genes during Adult Intestinal Organogenesis
Abstract
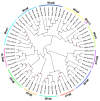
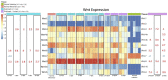
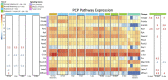
Wnt signaling has been shown to play multiple roles in regenerative processes, one of the most widely studied of which is the regeneration of the intestinal luminal epithelia. Most studies in this area have focused on self-renewal of the luminal stem cells; however, Wnt signaling may also have more dynamic functions, such as facilitating intestinal organogenesis. To explore this possibility, we employed the sea cucumber Holothuria glaberrima that can regenerate a full intestine over the course of 21 days after evisceration. We collected RNA-seq data from various intestinal tissues and regeneration stages and used these data to define the Wnt genes present in H. glaberrima and the differential gene expression (DGE) patterns during the regenerative process. Twelve Wnt genes were found, and their presence was confirmed in the draft genome of H. glaberrima. The expressions of additional Wnt-associated genes, such as Frizzled and Disheveled, as well as genes from the Wnt/β-catenin and Wnt/Planar Cell Polarity (PCP) pathways, were also analyzed. DGE showed unique distributions of Wnt in early- and late-stage intestinal regenerates, consistent with the Wnt/β-catenin pathway being upregulated during early-stages and the Wnt/PCP pathway being upregulated during late-stages. Our results demonstrate the diversity of Wnt signaling during intestinal regeneration, highlighting possible roles in adult organogenesis.
Keywords: Wnt genes; echinoderm; organogenesis; regeneration; sea cucumber.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Insights into intestinal regeneration signaling mechanisms.Dev Biol. 2020 Feb 1;458(1):12-31. doi: 10.1016/j.ydbio.2019.10.005. Epub 2019 Oct 9. Dev Biol. 2020. PMID: 31605680 Free PMC article.
-
Wnt/β-catenin signaling pathway regulates cell proliferation but not muscle dedifferentiation nor apoptosis during sea cucumber intestinal regeneration.Dev Biol. 2021 Dec;480:105-113. doi: 10.1016/j.ydbio.2021.08.011. Epub 2021 Sep 3. Dev Biol. 2021. PMID: 34481794 Free PMC article.
-
A Dynamic WNT/β-CATENIN Signaling Environment Leads to WNT-Independent and WNT-Dependent Proliferation of Embryonic Intestinal Progenitor Cells.Stem Cell Reports. 2016 Nov 8;7(5):826-839. doi: 10.1016/j.stemcr.2016.09.004. Epub 2016 Oct 6. Stem Cell Reports. 2016. PMID: 27720905 Free PMC article.
-
Targeting the Wnt/beta-catenin pathway in cancer: Update on effectors and inhibitors.Cancer Treat Rev. 2018 Jan;62:50-60. doi: 10.1016/j.ctrv.2017.11.002. Epub 2017 Nov 13. Cancer Treat Rev. 2018. PMID: 29169144 Free PMC article. Review.
-
The roles of Wnt/β-catenin pathway in tissue development and regenerative medicine.J Cell Physiol. 2018 Aug;233(8):5598-5612. doi: 10.1002/jcp.26265. Epub 2018 Mar 7. J Cell Physiol. 2018. PMID: 29150936 Review.
Cited by
-
A Novel Approach to Comparative RNA-Seq Does Not Support a Conserved Set of Orthologs Underlying Animal Regeneration.Genome Biol Evol. 2024 Jun 4;16(6):evae120. doi: 10.1093/gbe/evae120. Genome Biol Evol. 2024. PMID: 38922665 Free PMC article.
-
Single-cell RNA sequencing of the holothurian regenerating intestine reveals the pluripotency of the coelomic epithelium.Elife. 2025 Mar 20;13:RP100796. doi: 10.7554/eLife.100796. Elife. 2025. PMID: 40111904 Free PMC article.
-
Single-cell RNA sequencing of the holothurian regenerating intestine reveals the pluripotency of the coelomic epithelium.bioRxiv [Preprint]. 2024 Dec 31:2024.07.01.601561. doi: 10.1101/2024.07.01.601561. bioRxiv. 2024. Update in: Elife. 2025 Mar 20;13:RP100796. doi: 10.7554/eLife.100796. PMID: 39005414 Free PMC article. Updated. Preprint.
-
Evidence of interactions among apoptosis, cell proliferation, and dedifferentiation in the rudiment during whole-organ intestinal regeneration in the sea cucumber.Dev Biol. 2024 Jan;505:99-109. doi: 10.1016/j.ydbio.2023.11.001. Epub 2023 Nov 3. Dev Biol. 2024. PMID: 37925124 Free PMC article.
References
-
- Yaglova N.V., Tsomartova D.A., Obernikhin S.S., Nazimova S.V. The Role of the Canonical Wnt-Signaling Pathway in Morphogenesis and Regeneration of the Adrenal Cortex in Rats Exposed to the Endocrine Disruptor Dichlorodiphenyltrichloroethane during Prenatal and Postnatal Development. Biol. Bull. Russ. Acad. Sci. 2019;46:74–81. doi: 10.1134/S1062359018060122. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

