Excluding Digenic Inheritance of PGAP2 and PGAP3 Variants in Mabry Syndrome (OMIM 239300) Patient: Phenotypic Spectrum Associated with PGAP2 Gene Variants in Hyperphosphatasia with Mental Retardation Syndrome-3 (HPMRS3)
- PMID: 36833286
- PMCID: PMC9957281
- DOI: 10.3390/genes14020359
Excluding Digenic Inheritance of PGAP2 and PGAP3 Variants in Mabry Syndrome (OMIM 239300) Patient: Phenotypic Spectrum Associated with PGAP2 Gene Variants in Hyperphosphatasia with Mental Retardation Syndrome-3 (HPMRS3)
Abstract
We present a case report of a child with features of hyperphosphatasia with neurologic deficit (HPMRS) or Mabry syndrome (MIM 239300) with variants of unknown significance in two post-GPI attachments to proteins genes, PGAP2 and PGAP3, that underlie HPMRS 3 and 4.
Background: In addition to HPMRS 3 and 4, disruption of four phosphatidylinositol glycan (PIG) biosynthesis genes, PIGV, PIGO, PIGW and PIGY, result in HPMRS 1, 2, 5 and 6, respectively.
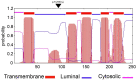
Methods: Targeted exome panel sequencing identified homozygous variants of unknown significance (VUS) in PGAP2 c:284A>G and PGAP3 c:259G>A. To assay the pathogenicity of these variants, we conducted a rescue assay in PGAP2 and PGAP3 deficient CHO cell lines.
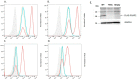
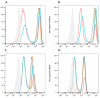
Results: Using a strong (pME) promoter, the PGAP2 variant did not rescue activity in CHO cells and the protein was not detected. Flow cytometric analysis showed that CD59 and CD55 expression on the PGAP2 deficient cell line was not restored by variant PGAP2. By contrast, activity of the PGAP3 variant was similar to wild-type.
Conclusions: For this patient with Mabry syndrome, the phenotype is likely to be predominantly HPMRS3: resulting from autosomal recessive inheritance of NM_001256240.2 PGAP2 c:284A>G, p.Tyr95Cys. We discuss strategies for establishing evidence for putative digenic inheritance in GPI deficiency disorders.
Keywords: CD55; CD59; Mabry syndrome; developmental disability; exome panel sequencing; flow cytometry; glycosylphosphatidylinositol (GPI) disorder; hyperphosphatasia with mental retardation (HPMRS); post attachment to proteins; rescue assay; type 2 (PGAP2).
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Thompson M.D., Nezarati M.M., Gillessen-Kaesbach G., Meinecke P., Mendoza R., Mornet E., Brun-Heath I., Squarcioni C.P., Legea-Mallet L., Munnich A., et al. Hyperphosphatasia with seizures, neurologic deficit, and characteristic facial features: Five new patients with Mabry syndrome. Am. J. Med. Genet. 2010;152A:1661–1669. - PubMed
-
- Cole D.E., Thompson M.D. Neurogenetic aspects of hyperphosphatasia in Mabry syndrome. Subcell. Biochem. 2015;76:343–361. - PubMed
-
- Carmody L.C., Blau H., Danis D., Zhang X.A., Gourdine J.-P., Vasilevsky N., Krawitz P., Thompson M.D., Robinson P.N. Significantly different clinical phenotypes associated with mutations in synthesis and transamidase + remodeling glycosylphosphatidylinositol (GPI)-anchor biosynthesis genes. Orphanet J. Rare Dis. 2020;15:40. doi: 10.1186/s13023-020-1313-0. - DOI - PMC - PubMed
-
- Krawitz P., Schweiger M.R., Rödelsperger C., Marcelis C., Kölsch U., Meisel C., Stephani F., Kinoshita T., Murakami Y., Bauer S., et al. Identity-by-descent filtering of exome sequence data identifies PIGV mutations in hyperphosphatasia mental retardation syndrome. Nat. Genet. 2010;42:827–829. doi: 10.1038/ng.653. - DOI - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

