Molecular Evolution of SARS-CoV-2 during the COVID-19 Pandemic
- PMID: 36833334
- PMCID: PMC9956206
- DOI: 10.3390/genes14020407
Molecular Evolution of SARS-CoV-2 during the COVID-19 Pandemic
Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) produced diverse molecular variants during its recent expansion in humans that caused different transmissibility and severity of the associated disease as well as resistance to monoclonal antibodies and polyclonal sera, among other treatments. In order to understand the causes and consequences of the observed SARS-CoV-2 molecular diversity, a variety of recent studies investigated the molecular evolution of this virus during its expansion in humans. In general, this virus evolves with a moderate rate of evolution, in the order of 10-3-10-4 substitutions per site and per year, which presents continuous fluctuations over time. Despite its origin being frequently associated with recombination events between related coronaviruses, little evidence of recombination was detected, and it was mostly located in the spike coding region. Molecular adaptation is heterogeneous among SARS-CoV-2 genes. Although most of the genes evolved under purifying selection, several genes showed genetic signatures of diversifying selection, including a number of positively selected sites that affect proteins relevant for the virus replication. Here, we review current knowledge about the molecular evolution of SARS-CoV-2 in humans, including the emergence and establishment of variants of concern. We also clarify relationships between the nomenclatures of SARS-CoV-2 lineages. We conclude that the molecular evolution of this virus should be monitored over time for predicting relevant phenotypic consequences and designing future efficient treatments.
Keywords: SARS-CoV-2; genetic diversity; molecular adaptation; molecular evolution; rate of evolution; recombination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
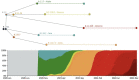
References
-
- Bonotti M., Zech S.T. Recovering Civility during COVID-19. Springer; Berlin/Heidelberg, Germany: 2021. The Human, Economic, Social, and Political Costs of COVID-19; pp. 1–36. - DOI
-
- Pan T., Chen R., He X., Yuan Y., Deng X., Li R., Yan H., Yan S., Liu J., Zhang Y., et al. Infection of wild-type mice by SARS-CoV-2 B. 1.351 variant indicates a possible novel cross-species transmission route. Signal Transduct. Target. Ther. 2021;6:420. doi: 10.1038/s41392-021-00848-1. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

