The Identification and Characteristics of miRNAs Related to Cashmere Fiber Traits in Skin Tissue of Cashmere Goats
- PMID: 36833400
- PMCID: PMC9957446
- DOI: 10.3390/genes14020473
The Identification and Characteristics of miRNAs Related to Cashmere Fiber Traits in Skin Tissue of Cashmere Goats
Abstract
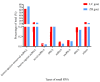
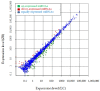
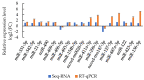
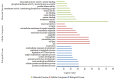
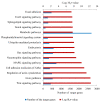
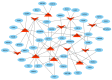
microRNAs (miRNAs) are involved in the regulation of biological phenomena by down-regulating the expression of mRNAs. In this study, Liaoning cashmere (LC) goats (n = 6) and Ziwuling black (ZB) goats (n = 6) with different cashmere fiber production performances were selected. We supposed that miRNAs are responsible for the cashmere fiber trait differences. To test the hypothesis, the expression profiles of miRNAs from the skin tissue of the two caprine breeds were compared using small RNA sequencing (RNA-seq). A total of 1293 miRNAs were expressed in the caprine skin samples, including 399 known caprine miRNAs, 691 known species-conserved miRNAs, and 203 novel miRNAs. Compared with ZB goats, 112 up-regulated miRNAs, and 32 down-regulated miRNAs were found in LC goats. The target genes of the differentially expressed miRNAs were remarkably concentrated on some terms and pathways associated with cashmere fiber performance, including binding, cell, cellular protein modification process, and Wnt, Notch, and MAPK signaling pathways. The miRNA-mRNA interaction network found that 14 miRNAs selected may contribute to cashmere fiber traits regulation by targeting functional genes associated with hair follicle activities. The results have reinforced others leading to a solid foundation for further investigation of the influences of individual miRNAs on cashmere fiber traits in cashmere goats.
Keywords: cashmere fiber properties; cashmere goats; miRNAs; small RNA sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Deciphering the molecular drivers for cashmere/pashmina fiber production in goats: a comprehensive review.Mamm Genome. 2025 Mar;36(1):162-182. doi: 10.1007/s00335-025-10109-z. Epub 2025 Feb 4. Mamm Genome. 2025. PMID: 39904908 Review.
-
RNA-Seq Reveals the Roles of Long Non-Coding RNAs (lncRNAs) in Cashmere Fiber Production Performance of Cashmere Goats in China.Genes (Basel). 2023 Feb 1;14(2):384. doi: 10.3390/genes14020384. Genes (Basel). 2023. PMID: 36833312 Free PMC article.
-
Identification and Characterization of Circular RNAs (circRNAs) Using RNA-Seq in Two Breeds of Cashmere Goats.Genes (Basel). 2023 Jan 27;14(2):331. doi: 10.3390/genes14020331. Genes (Basel). 2023. PMID: 36833256 Free PMC article.
-
Regulation of secondary hair follicle cycle in cashmere goats by miR-877-3p targeting IGFBP5 gene.J Anim Sci. 2023 Jan 3;101:skad314. doi: 10.1093/jas/skad314. J Anim Sci. 2023. PMID: 37777862 Free PMC article.
-
Integrative analysis reveals ncRNA-mediated molecular regulatory network driving secondary hair follicle regression in cashmere goats.BMC Genomics. 2018 Mar 27;19(1):222. doi: 10.1186/s12864-018-4603-3. BMC Genomics. 2018. PMID: 29587631 Free PMC article.
Cited by
-
Multi-omics and AI-driven advances in miRNA-mediated hair follicle regulation in cashmere goats.Front Vet Sci. 2025 Jul 9;12:1635202. doi: 10.3389/fvets.2025.1635202. eCollection 2025. Front Vet Sci. 2025. PMID: 40703926 Free PMC article. Review.
-
Molecular insights into Pashmina fiber production: comparative skin transcriptomic analysis of Changthangi goats and sheep.Mamm Genome. 2024 Jun;35(2):160-169. doi: 10.1007/s00335-024-10040-9. Epub 2024 Apr 8. Mamm Genome. 2024. PMID: 38589518
-
Deciphering the molecular drivers for cashmere/pashmina fiber production in goats: a comprehensive review.Mamm Genome. 2025 Mar;36(1):162-182. doi: 10.1007/s00335-025-10109-z. Epub 2025 Feb 4. Mamm Genome. 2025. PMID: 39904908 Review.
References
-
- Waldron S.A., Komarek A.M., Brown C.G. An exploration of livestock-development policies in western China. Food Policy. 2012;37:12–20.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

