The Non-JAZ TIFY Protein TIFY8 of Arabidopsis thaliana Interacts with the HD-ZIP III Transcription Factor REVOLUTA and Regulates Leaf Senescence
- PMID: 36834490
- PMCID: PMC9967580
- DOI: 10.3390/ijms24043079
The Non-JAZ TIFY Protein TIFY8 of Arabidopsis thaliana Interacts with the HD-ZIP III Transcription Factor REVOLUTA and Regulates Leaf Senescence
Abstract
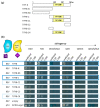
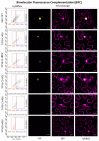
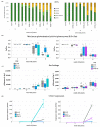
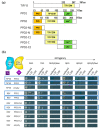
The HD-ZIP III transcription factor REVOLUTA (REV) is involved in early leaf development, as well as in leaf senescence. REV directly binds to the promoters of senescence-associated genes, including the central regulator WRKY53. As this direct regulation appears to be restricted to senescence, we aimed to characterize protein-interaction partners of REV which could mediate this senescence-specificity. The interaction between REV and the TIFY family member TIFY8 was confirmed by yeast two-hybrid assays, as well as by bimolecular fluorescence complementation in planta. This interaction inhibited REV's function as an activator of WRKY53 expression. Mutation or overexpression of TIFY8 accelerated or delayed senescence, respectively, but did not significantly alter early leaf development. Jasmonic acid (JA) had only a limited effect on TIFY8 expression or function; however, REV appears to be under the control of JA signaling. Accordingly, REV also interacted with many other members of the TIFY family, namely the PEAPODs and several JAZ proteins in the yeast system, which could potentially mediate the JA-response. Therefore, REV appears to be under the control of the TIFY family in two different ways: a JA-independent way through TIFY8, which controls REV function in senescence, and a JA-dependent way through PEAPODs and JAZ proteins.
Keywords: Arabidopsis thaliana; JAZ proteins; PEAPOD; REVOLUTA; TIFY8; jasmonic acid signaling; leaf senescence; transcription factor regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
The non-JAZ TIFY protein TIFY8 from Arabidopsis thaliana is a transcriptional repressor.PLoS One. 2014 Jan 8;9(1):e84891. doi: 10.1371/journal.pone.0084891. eCollection 2014. PLoS One. 2014. PMID: 24416306 Free PMC article.
-
REVOLUTA and WRKY53 connect early and late leaf development in Arabidopsis.Development. 2014 Dec;141(24):4772-83. doi: 10.1242/dev.117689. Epub 2014 Nov 13. Development. 2014. PMID: 25395454 Free PMC article.
-
MYB47 delays leaf senescence by modulating jasmonate pathway via direct regulation of CYP94B3/CYP94C1 expression in Arabidopsis.New Phytol. 2025 Jun;246(5):2192-2206. doi: 10.1111/nph.70133. Epub 2025 Apr 5. New Phytol. 2025. PMID: 40186431
-
Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones.J Exp Bot. 2017 Mar 1;68(6):1361-1369. doi: 10.1093/jxb/erx004. J Exp Bot. 2017. PMID: 28201612 Review.
-
JAZ repressors and the orchestration of phytohormone crosstalk.Trends Plant Sci. 2012 Jan;17(1):22-31. doi: 10.1016/j.tplants.2011.10.006. Epub 2011 Nov 21. Trends Plant Sci. 2012. PMID: 22112386 Review.
Cited by
-
Genome-wide identification and expression analysis of TIFY genes under MeJA, cold and PEG-induced drought stress treatment in Dendrobium huoshanense.Physiol Mol Biol Plants. 2024 Apr;30(4):527-542. doi: 10.1007/s12298-024-01442-9. Epub 2024 Mar 25. Physiol Mol Biol Plants. 2024. PMID: 38737319 Free PMC article.
-
Regulatory Mechanisms of Transcription Factors in Plant Morphology and Function.Int J Mol Sci. 2023 Apr 11;24(8):7039. doi: 10.3390/ijms24087039. Int J Mol Sci. 2023. PMID: 37108201 Free PMC article.
-
Genome-wide identification, characterization and expression pattern analysis of TIFY family members in Artemisia argyi.BMC Genomics. 2024 Oct 3;25(1):925. doi: 10.1186/s12864-024-10856-4. BMC Genomics. 2024. PMID: 39363209 Free PMC article.
-
Photosynthesis Response and Transcriptional Analysis: Dissecting the Role of SlHB8 in Regulating Drought Resistance in Tomato Plants.Int J Mol Sci. 2023 Oct 23;24(20):15498. doi: 10.3390/ijms242015498. Int J Mol Sci. 2023. PMID: 37895176 Free PMC article.
-
Genome-Wide Identification and Expressional Analysis of the TIFY Gene Family in Eucalyptus grandis.Int J Mol Sci. 2025 Aug 16;26(16):7914. doi: 10.3390/ijms26167914. Int J Mol Sci. 2025. PMID: 40869233 Free PMC article.
References
-
- Hong S.Y., Botterweg-Paredes E., Doll J., Eguen T., Blaakmeer A., Matton S., Xie Y., Skjøth Lunding B., Zentgraf U., Guan C., et al. Multi-level analysis of the interactions between REVOLUTA and MORE AXILLARY BRANCHES 2 in controlling plant development reveals parallel, independent and antagonistic functions. Development. 2020;14:dev183681. doi: 10.1242/dev.183681. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

