Whole Transcriptome Analysis of Hypothalamus in Mice during Short-Term Starvation
- PMID: 36834616
- PMCID: PMC9968171
- DOI: 10.3390/ijms24043204
Whole Transcriptome Analysis of Hypothalamus in Mice during Short-Term Starvation
Abstract
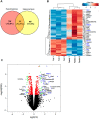
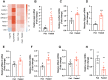
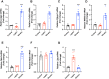
Molecular profiling of the hypothalamus in response to metabolic shifts is a critical cue to better understand the principle of the central control of whole-body energy metabolism. The transcriptional responses of the rodent hypothalamus to short-term calorie restriction have been documented. However, studies on the identification of hypothalamic secretory factors that potentially contribute to the control of appetite are lacking. In this study, we analyzed the differential expression of hypothalamic genes and compared the selected secretory factors from the fasted mice with those of fed control mice using bulk RNA-sequencing. We verified seven secretory genes that were significantly altered in the hypothalamus of fasted mice. In addition, we determined the response of secretory genes in cultured hypothalamic cells to treatment with ghrelin and leptin. The current study provides further insights into the neuronal response to food restriction at the molecular level and may be useful for understanding the hypothalamic control of appetite.
Keywords: RNA-seq; appetite; energy homeostasis; food deprivation; hypothalamus; secretory factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Nestor C.C., Qiu J., Padilla S.L., Zhang C., Bosch M.A., Fan W., Aicher S.A., Palmiter R.D., Rønnekleiv O.K., Kelly M.J. Optogenetic Stimulation of Arcuate Nucleus Kiss1 Neurons Reveals a Steroid-Dependent Glutamatergic Input to POMC and AgRP Neurons in Male Mice. Mol. Endocrinol. 2016;30:630–644. doi: 10.1210/me.2016-1026. - DOI - PMC - PubMed
-
- Yu S., Qualls-Creekmore E., Rezai-Zadeh K., Jiang Y., Berthoud H.-R., Morrison C.D., Derbenev A.V., Zsombok A., Münzberg H. Glutamatergic Preoptic Area Neurons That Express Leptin Receptors Drive Temperature-Dependent Body Weight Homeostasis. J. Neurosci. 2016;36:5034–5046. doi: 10.1523/JNEUROSCI.0213-16.2016. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

