Olive Polyphenol Oxidase Gene Family
- PMID: 36834644
- PMCID: PMC9962951
- DOI: 10.3390/ijms24043233
Olive Polyphenol Oxidase Gene Family
Abstract
The phenolic compounds containing hydroxytyrosol are the minor components of virgin olive oil (VOO) with the greatest impact on its functional properties and health benefits. Olive breeding for improving the phenolic composition of VOO is strongly dependent on the identification of the key genes determining the biosynthesis of these compounds in the olive fruit and also their transformation during the oil extraction process. In this work, olive polyphenol oxidase (PPO) genes have been identified and fully characterized in order to evaluate their specific role in the metabolism of hydroxytyrosol-derived compounds by combining gene expression analysis and metabolomics data. Four PPO genes have been identified, synthesized, cloned and expressed in Escherichia coli, and the functional identity of the recombinant proteins has been verified using olive phenolic substrates. Among the characterized genes, two stand out: (i) OePPO2 with its diphenolase activity, which is very active in the oxidative degradation of phenols during oil extraction and also seems to be highly involved in the natural defense mechanism in response to biotic stress, and (ii) OePPO3, which codes for a tyrosinase protein, having diphenolase but also monophenolase activity, which catalyzes the hydroxylation of tyrosol to form hydroxytyrosol.
Keywords: Olea europaea L.; hydroxytyrosol; phenolic metabolism; polyphenol oxidase; tyrosinase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





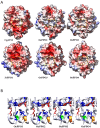

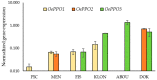

References
-
- Parra-Pérez A.M., Pérez-Jiménez A., Gris-Cárdenas I., Bonel-Pérez G.C., Carrasco-Díaz L.M., Mokhtari K., García-Salguero L., Lupiáñez J.A., Rufino-Palomares E.E. Involvement of the PI3K/AKT intracellular signaling pathway in the anticancer activity of hydroxytyrosol, a polyphenol from Olea europaea, in hematological cells and implication of HSP60 levels in its anti-inflamatory activity. Int. J. Mol. Sci. 2022;23:7053. doi: 10.3390/ijms23137053. - DOI - PMC - PubMed
-
- Laghezza Masci V., Bernini R., Villanova N., Clemente M., Cicaloni V., Tinti L., Salvini L., Taddei A.R., Tiezzi A., Ovidi E. In vitro anti-poliferative and apoptotic effects of hydroxytyrosyl oleate on SH-SY5Y human neuroblastoma cells. Int. J. Mol. Sci. 2022;23:12348. doi: 10.3390/ijms232012348. - DOI - PMC - PubMed
-
- European Commission Commission Regulation (EU) No 1018/2013 amending Regulation (EU) No 432/2012 establishing a list of permitted health claims made on foods other than those referring to the reduction of disease risk and to children’s development and heal. Off. J. Eur. Union. 2012;282:43–45.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

