The Chromatin Landscape around DNA Double-Strand Breaks in Yeast and Its Influence on DNA Repair Pathway Choice
- PMID: 36834658
- PMCID: PMC9967470
- DOI: 10.3390/ijms24043248
The Chromatin Landscape around DNA Double-Strand Breaks in Yeast and Its Influence on DNA Repair Pathway Choice
Abstract
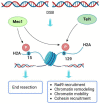
DNA double-strand breaks (DSBs) are harmful DNA lesions, which elicit catastrophic consequences for genome stability if not properly repaired. DSBs can be repaired by either non-homologous end joining (NHEJ) or homologous recombination (HR). The choice between these two pathways depends on which proteins bind to the DSB ends and how their action is regulated. NHEJ initiates with the binding of the Ku complex to the DNA ends, while HR is initiated by the nucleolytic degradation of the 5'-ended DNA strands, which requires several DNA nucleases/helicases and generates single-stranded DNA overhangs. DSB repair occurs within a precisely organized chromatin environment, where the DNA is wrapped around histone octamers to form the nucleosomes. Nucleosomes impose a barrier to the DNA end processing and repair machinery. Chromatin organization around a DSB is modified to allow proper DSB repair either by the removal of entire nucleosomes, thanks to the action of chromatin remodeling factors, or by post-translational modifications of histones, thus increasing chromatin flexibility and the accessibility of repair enzymes to the DNA. Here, we review histone post-translational modifications occurring around a DSB in the yeast Saccharomyces cerevisiae and their role in DSB repair, with particular attention to DSB repair pathway choice.
Keywords: DNA double strand break; NHEJ; Saccharomyces cerevisiae; histone acetylation; histone methylation; histone phosphorylation; histone ubiquitylation; homologous recombination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

