Dependence on MUC1-C in Progression of Neuroendocrine Prostate Cancer
- PMID: 36835130
- PMCID: PMC9967814
- DOI: 10.3390/ijms24043719
Dependence on MUC1-C in Progression of Neuroendocrine Prostate Cancer
Abstract
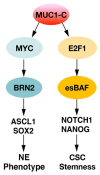
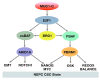
Castration resistant prostate cancer (CRPC) is responsive to androgen receptor (AR) axis targeted agents; however, patients invariably relapse with resistant disease that often progresses to neuroendocrine prostate cancer (NEPC). Treatment-related NEPC (t-NEPC) is highly aggressive with limited therapeutic options and poor survival outcomes. The molecular basis for NEPC progression remains incompletely understood. The MUC1 gene evolved in mammals to protect barrier tissues from loss of homeostasis. MUC1 encodes the transmembrane MUC1-C subunit, which is activated by inflammation and contributes to wound repair. However, chronic activation of MUC1-C contributes to lineage plasticity and carcinogenesis. Studies in human NEPC cell models have demonstrated that MUC1-C suppresses the AR axis and induces the Yamanaka OSKM pluripotency factors. MUC1-C interacts directly with MYC and activates the expression of the BRN2 neural transcription factor (TF) and other effectors, such as ASCL1, of the NE phenotype. MUC1-C also induces the NOTCH1 stemness TF in promoting the NEPC cancer stem cell (CSC) state. These MUC1-C-driven pathways are coupled with activation of the SWI/SNF embryonic stem BAF (esBAF) and polybromo-BAF (PBAF) chromatin remodeling complexes and global changes in chromatin architecture. The effects of MUC1-C on chromatin accessibility integrate the CSC state with the control of redox balance and induction of self-renewal capacity. Importantly, targeting MUC1-C inhibits NEPC self-renewal, tumorigenicity and therapeutic resistance. This dependence on MUC1-C extends to other NE carcinomas, such as SCLC and MCC, and identify MUC1-C as a target for the treatment of these aggressive malignancies with the anti-MUC1 agents now under clinical and preclinical development.
Keywords: CSC; MUC1-C; NEPC; chromatin remodeling; lineage plasticity.
Conflict of interest statement
D.K. has equity interests in Reata Pharmaceuticals and Hillstream Biopharma and is a paid consultant to Reata and CanBas.
Figures

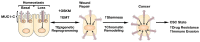

Similar articles
-
MUC1-C regulates lineage plasticity driving progression to neuroendocrine prostate cancer.Nat Commun. 2020 Jan 17;11(1):338. doi: 10.1038/s41467-019-14219-6. Nat Commun. 2020. PMID: 31953400 Free PMC article.
-
MUC1-C Activates the BAF (mSWI/SNF) Complex in Prostate Cancer Stem Cells.Cancer Res. 2021 Feb 15;81(4):1111-1122. doi: 10.1158/0008-5472.CAN-20-2588. Epub 2020 Dec 15. Cancer Res. 2021. PMID: 33323379 Free PMC article.
-
MUC1-C dependence in treatment-resistant prostate cancer uncovers a target for antibody-drug conjugate therapy.JCI Insight. 2025 Jun 24;10(14):e190924. doi: 10.1172/jci.insight.190924. eCollection 2025 Jul 22. JCI Insight. 2025. PMID: 40553569 Free PMC article.
-
Clinical and Biological Features of Neuroendocrine Prostate Cancer.Curr Oncol Rep. 2021 Jan 12;23(2):15. doi: 10.1007/s11912-020-01003-9. Curr Oncol Rep. 2021. PMID: 33433737 Free PMC article. Review.
-
Addiction of Cancer Stem Cells to MUC1-C in Triple-Negative Breast Cancer Progression.Int J Mol Sci. 2022 Jul 26;23(15):8219. doi: 10.3390/ijms23158219. Int J Mol Sci. 2022. PMID: 35897789 Free PMC article. Review.
Cited by
-
NCAPG2 promotes prostate cancer malignancy and stemness via STAT3/c-MYC signaling.J Transl Med. 2024 Jan 2;22(1):12. doi: 10.1186/s12967-023-04834-9. J Transl Med. 2024. PMID: 38166947 Free PMC article.
-
MUC1-C is a target of salinomycin in inducing ferroptosis of cancer stem cells.Cell Death Discov. 2024 Jan 5;10(1):9. doi: 10.1038/s41420-023-01772-9. Cell Death Discov. 2024. PMID: 38182558 Free PMC article.
-
The Role of MUC1 in Renal Cell Carcinoma.Biomolecules. 2024 Mar 7;14(3):315. doi: 10.3390/biom14030315. Biomolecules. 2024. PMID: 38540735 Free PMC article. Review.
-
New Insights into Potential Therapeutic Targets for Neuroendocrine Prostate Cancer: From Bench to Clinic.Research (Wash D C). 2025 Jul 31;8:0791. doi: 10.34133/research.0791. eCollection 2025. Research (Wash D C). 2025. PMID: 40746825 Free PMC article. Review.
-
MUC1-C auto-regulatory complex with EBNA1 is responsible for latent Epstein-Barr virus-associated gastric cancer progression.Oncogene. 2025 Aug 5. doi: 10.1038/s41388-025-03519-5. Online ahead of print. Oncogene. 2025. PMID: 40764753
References
-
- Aggarwal R., Huang J., Alumkal J.J., Zhang L., Feng F.Y., Thomas G.V., Weinstein A.S., Friedl V., Zhang C., Witte O.N., et al. Clinical and Genomic Characterization of Treatment-Emergent Small-Cell Neuroendocrine Prostate Cancer: A Multi-institutional Prospective Study. J. Clin. Oncol. 2018;36:2492–2503. doi: 10.1200/JCO.2017.77.6880. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

