Detection of Breast Cancer-Specific Extracellular Vesicles with Fiber-Optic SPR Biosensor
- PMID: 36835174
- PMCID: PMC9966403
- DOI: 10.3390/ijms24043764
Detection of Breast Cancer-Specific Extracellular Vesicles with Fiber-Optic SPR Biosensor
Abstract
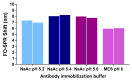
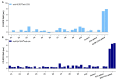
Extracellular vesicles (EVs) have attracted great attention as potential biomarkers for cancer diagnostics. Although several technologies have been developed for EV detection, many of them are still not applicable to clinical settings as they rely on complex EV isolation processes, while lacking sensitivity, specificity or standardization. To solve this problem, we have developed a sensitive breast cancer-specific EV detection bioassay directly in blood plasma using a fiber-optic surface plasmon resonance (FO-SPR) biosensor, previously calibrated with recombinant EVs. First, we established a sandwich bioassay to detect SK-BR-3 EVs by functionalizing the FO-SPR probes with anti-HER2 antibodies. A calibration curve was built using an anti-HER2/Banti-CD9 combination, resulting in an LOD of 2.1 × 107 particles/mL in buffer and 7 × 108 particles/mL in blood plasma. Next, we investigated the potential of the bioassay to detect MCF7 EVs in blood plasma using an anti-EpCAM/Banti-mix combination, obtaining an LOD of 1.1 × 10 8 particles/mL. Finally, the specificity of the bioassay was proven by the absence of signal when testing plasma samples from 10 healthy people unknown to be diagnosed with breast cancer. The remarkable sensitivity and specificity of the developed sandwich bioassay together with the advantages of the standardized FO-SPR biosensor highlight outstanding potential for the future of EV analysis.
Keywords: EpCAM; HER2; biosensors; breast cancer; extracellular vesicles; fiber-optic surface plasmon resonance.
Conflict of interest statement
Jeroen Lammertyn is a board member of FOx Biosystems, a spin-off company of KU Leuven commercializing FO-SPR platforms, next to the principal investigator of the Biosensors group.
Figures




References
-
- Simeone P., Bologna G., Lanuti P., Pierdomenico L., Guagnano M.T., Pieragostino D., del Boccio P., Vergara D., Marchisio M., Miscia S., et al. Extracellular Vesicles as Signaling Mediators and Disease Biomarkers across Biological Barriers. Int. J. Mol. Sci. 2020;21:2514. doi: 10.3390/ijms21072514. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

