Comparison of Tomato Transcriptomic Profiles Reveals Overlapping Patterns in Abiotic and Biotic Stress Responses
- PMID: 36835470
- PMCID: PMC9961515
- DOI: 10.3390/ijms24044061
Comparison of Tomato Transcriptomic Profiles Reveals Overlapping Patterns in Abiotic and Biotic Stress Responses
Abstract
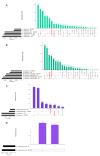
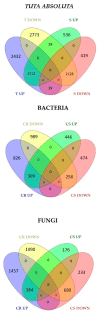
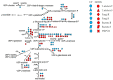
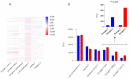
Until a few years ago, many studies focused on the transcriptomic response to single stresses. However, tomato cultivations are often constrained by a wide range of biotic and abiotic stress that can occur singularly or in combination, and several genes can be involved in the defensive mechanism response. Therefore, we analyzed and compared the transcriptomic responses of resistant and susceptible genotypes to seven biotic stresses (Cladosporium fulvum, Phytophthora infestans, Pseudomonas syringae, Ralstonia solanacearum, Sclerotinia sclerotiorum, Tomato spotted wilt virus (TSWV) and Tuta absoluta) and five abiotic stresses (drought, salinity, low temperatures, and oxidative stress) to identify genes involved in response to multiple stressors. With this approach, we found genes encoding for TFs, phytohormones, or participating in signaling and cell wall metabolic processes, participating in defense against various biotic and abiotic stress. Moreover, a total of 1474 DEGs were commonly found between biotic and abiotic stress. Among these, 67 DEGs were involved in response to at least four different stresses. In particular, we found RLKs, MAPKs, Fasciclin-like arabinogalactans (FLAs), glycosyltransferases, genes involved in the auxin, ET, and JA pathways, MYBs, bZIPs, WRKYs and ERFs genes. Detected genes responsive to multiple stress might be further investigated with biotechnological approaches to effectively improve plant tolerance in the field.
Keywords: cell wall; genome editing; phytohormones; plant stress; signaling; transcription factors; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Transcriptome Analysis Reveals New Insights into the Bacterial Wilt Resistance Mechanism Mediated by Silicon in Tomato.Int J Mol Sci. 2019 Feb 11;20(3):761. doi: 10.3390/ijms20030761. Int J Mol Sci. 2019. PMID: 30754671 Free PMC article.
-
Auxin Response Factors (ARFs) are potential mediators of auxin action in tomato response to biotic and abiotic stress (Solanum lycopersicum).PLoS One. 2018 Feb 28;13(2):e0193517. doi: 10.1371/journal.pone.0193517. eCollection 2018. PLoS One. 2018. PMID: 29489914 Free PMC article.
-
The LEA gene family in tomato and its wild relatives: genome-wide identification, structural characterization, expression profiling, and role of SlLEA6 in drought stress.BMC Plant Biol. 2022 Dec 19;22(1):596. doi: 10.1186/s12870-022-03953-7. BMC Plant Biol. 2022. PMID: 36536303 Free PMC article.
-
Plant behaviour under combined stress: tomato responses to combined salinity and pathogen stress.Plant J. 2018 Feb;93(4):781-793. doi: 10.1111/tpj.13800. Epub 2018 Jan 19. Plant J. 2018. PMID: 29237240 Review.
-
Plant Immune System: Crosstalk Between Responses to Biotic and Abiotic Stresses the Missing Link in Understanding Plant Defence.Curr Issues Mol Biol. 2017;23:1-16. doi: 10.21775/cimb.023.001. Epub 2017 Feb 3. Curr Issues Mol Biol. 2017. PMID: 28154243 Review.
Cited by
-
Revitalizing agriculture: next-generation genotyping and -omics technologies enabling molecular prediction of resilient traits in the Solanaceae family.Front Plant Sci. 2024 Feb 5;15:1278760. doi: 10.3389/fpls.2024.1278760. eCollection 2024. Front Plant Sci. 2024. PMID: 38375087 Free PMC article. Review.
-
Tomato mitogen-activated protein kinase: mechanisms of adaptation in response to biotic and abiotic stresses.Front Plant Sci. 2025 Feb 3;16:1533248. doi: 10.3389/fpls.2025.1533248. eCollection 2025. Front Plant Sci. 2025. PMID: 39963529 Free PMC article. Review.
-
SEL1L3 as a link molecular between renal cell carcinoma and atherosclerosis based on bioinformatics analysis and experimental verification.Aging (Albany NY). 2023 Nov 21;15(22):13150-13162. doi: 10.18632/aging.205227. Epub 2023 Nov 21. Aging (Albany NY). 2023. PMID: 37993256 Free PMC article.
-
The First Genome-Wide Mildew Locus O Genes Characterization in the Lamiaceae Plant Family.Int J Mol Sci. 2023 Sep 4;24(17):13627. doi: 10.3390/ijms241713627. Int J Mol Sci. 2023. PMID: 37686433 Free PMC article.
-
Moss-pathogen interactions: a review of the current status and future opportunities.Front Genet. 2025 Feb 11;16:1539311. doi: 10.3389/fgene.2025.1539311. eCollection 2025. Front Genet. 2025. PMID: 40008229 Free PMC article. Review.
References
-
- Kesawat M.S., Kherawat B.S., Singh A., Dey P., Kabi M., Debnath D., Saha D., Khandual A., Rout S., Manorama, et al. Genome-Wide Identification and Characterization of the Brassinazole-Resistant (BZR) Gene Family and Its Expression in the Various Developmental Stage and Stress Conditions in Wheat (Triticum aestivum L.) Int. J. Mol. Sci. 2021;22:8743. doi: 10.3390/ijms22168743. - DOI - PMC - PubMed
-
- Kesawat M.S., Kherawat B.S., Singh A., Dey P., Routray S., Mohapatra C., Saha D., Ram C., Siddique K.H.M., Kumar A., et al. Genome-Wide Analysis and Characterization of the Proline-Rich Extensin-like Receptor Kinases (PERKs) Gene Family Reveals Their Role in Different Developmental Stages and Stress Conditions in Wheat (Triticum aestivum L.) Plants. 2022;11:496. doi: 10.3390/plants11040496. - DOI - PMC - PubMed
-
- Shao W., Chen W., Zhu X., Zhou X., Jin Y., Zhan C., Liu G., Liu X., Ma D., Qiao Y. Genome-Wide Identification and Characterization of Wheat 14-3-3 Genes Unravels the Role of Tagrf6-a in Salt Stress Tolerance by Binding Myb Transcription Factor. Int. J. Mol. Sci. 2021;22:1904. doi: 10.3390/ijms22041904. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

