Genome-Wide Identification, Classification, and Expression Analyses of the CsDGAT Gene Family in Cannabis sativa L. and Their Response to Cold Treatment
- PMID: 36835488
- PMCID: PMC9963917
- DOI: 10.3390/ijms24044078
Genome-Wide Identification, Classification, and Expression Analyses of the CsDGAT Gene Family in Cannabis sativa L. and Their Response to Cold Treatment
Abstract
Hempseed is a nutrient-rich natural resource, and high levels of hempseed oil accumulate within hemp seeds, consisting primarily of different triglycerides. Members of the diacylglycerol acyltransferase (DGAT) enzyme family play critical roles in catalyzing triacylglycerol biosynthesis in plants, often governing the rate-limiting step in this process. As such, this study was designed to characterize the Cannabis sativa DGAT (CsDGAT) gene family in detail. Genomic analyses of the C. sativa revealed 10 candidate DGAT genes that were classified into four families (DGAT1, DGAT2, DGAT3, WS/DGAT) based on the features of different isoforms. Members of the CsDGAT family were found to be associated with large numbers of cis-acting promoter elements, including plant response elements, plant hormone response elements, light response elements, and stress response elements, suggesting roles for these genes in key processes such as development, environmental adaptation, and abiotic stress responses. Profiling of these genes in various tissues and varieties revealed varying spatial patterns of CsDGAT expression dynamics and differences in expression among C. sativa varieties, suggesting that the members of this gene family likely play distinct functional regulatory functions CsDGAT genes were upregulated in response to cold stress, and significant differences in the mode of regulation were observed when comparing roots and leaves, indicating that CsDGAT genes may play positive roles as regulators of cold responses in hemp while also playing distinct roles in shaping the responses of different parts of hemp seedlings to cold exposure. These data provide a robust basis for further functional studies of this gene family, supporting future efforts to screen the significance of CsDGAT candidate genes to validate their functions to improve hempseed oil composition.
Keywords: diacylglycerol acyltransferase (DGAT); expression patterns; gene family; hemp.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

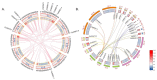


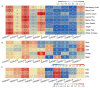


References
-
- Karche T., Singh M. The application of hemp (Cannabis sativa L.) for a green economy: A review. Turk. J. Bot. 2019;43:710–723. doi: 10.3906/bot-1907-15. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

