Gene Expression Profile as a Predictor of Seizure Liability
- PMID: 36835526
- PMCID: PMC9963992
- DOI: 10.3390/ijms24044116
Gene Expression Profile as a Predictor of Seizure Liability
Abstract
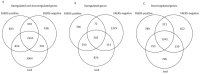
Analysis platforms to predict drug-induced seizure liability at an early phase of drug development would improve safety and reduce attrition and the high cost of drug development. We hypothesized that a drug-induced in vitro transcriptomics signature predicts its ictogenicity. We exposed rat cortical neuronal cultures to non-toxic concentrations of 34 compounds for 24 h; 11 were known to be ictogenic (tool compounds), 13 were associated with a high number of seizure-related adverse event reports in the clinical FDA Adverse Event Reporting System (FAERS) database and systematic literature search (FAERS-positive compounds), and 10 were known to be non-ictogenic (FAERS-negative compounds). The drug-induced gene expression profile was assessed from RNA-sequencing data. Transcriptomics profiles induced by the tool, FAERS-positive and FAERS-negative compounds, were compared using bioinformatics and machine learning. Of the 13 FAERS-positive compounds, 11 induced significant differential gene expression; 10 of the 11 showed an overall high similarity to the profile of at least one tool compound, correctly predicting the ictogenicity. Alikeness-% based on the number of the same differentially expressed genes correctly categorized 85%, the Gene Set Enrichment Analysis score correctly categorized 73%, and the machine-learning approach correctly categorized 91% of the FAERS-positive compounds with reported seizure liability currently in clinical use. Our data suggest that the drug-induced gene expression profile could be used as a predictive biomarker for seizure liability.
Keywords: attrition; cell culture; drug; gene expression; ictogenicity; neuronal.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Similar articles
-
Profiling cumulative proportional reporting ratios of drug-induced liver injury in the FDA Adverse Event Reporting System (FAERS) database.Drug Saf. 2013 Dec;36(12):1169-78. doi: 10.1007/s40264-013-0116-9. Drug Saf. 2013. PMID: 24178291
-
Complementing the US Food and Drug Administration Adverse Event Reporting System With Adverse Drug Reaction Reporting From Social Media: Comparative Analysis.JMIR Public Health Surveill. 2020 Sep 30;6(3):e19266. doi: 10.2196/19266. JMIR Public Health Surveill. 2020. PMID: 32996889 Free PMC article.
-
The Weber effect and the United States Food and Drug Administration's Adverse Event Reporting System (FAERS): analysis of sixty-two drugs approved from 2006 to 2010.Drug Saf. 2014 Apr;37(4):283-94. doi: 10.1007/s40264-014-0150-2. Drug Saf. 2014. PMID: 24643967 Free PMC article.
-
Emerging Causes of Drug-Induced Anaphylaxis: A Review of Anaphylaxis-Associated Reports in the FDA Adverse Event Reporting System (FAERS).J Allergy Clin Immunol Pract. 2021 Feb;9(2):819-829.e2. doi: 10.1016/j.jaip.2020.09.021. Epub 2020 Sep 28. J Allergy Clin Immunol Pract. 2021. PMID: 32992044 Free PMC article. Review.
-
Analysis of Spontaneous Postmarket Case Reports Submitted to the FDA Regarding Thromboembolic Adverse Events and JAK Inhibitors.Drug Saf. 2018 Apr;41(4):357-361. doi: 10.1007/s40264-017-0622-2. Drug Saf. 2018. PMID: 29196988 Review.
Cited by
-
Region-independent active CNS net uptake of marketed H+/OC antiporter system substrates.Front Cell Neurosci. 2024 Oct 29;18:1493644. doi: 10.3389/fncel.2024.1493644. eCollection 2024. Front Cell Neurosci. 2024. PMID: 39534684 Free PMC article.
-
Innovative Medicines Initiative public-private partnerships to enhance translational safety.Nat Rev Drug Discov. 2025 May 22. doi: 10.1038/d41573-025-00080-4. Online ahead of print. Nat Rev Drug Discov. 2025. PMID: 40404858 No abstract available.
References
-
- Authier S., Arezzo J., Delatte M.S., Kallman M.J., Markgraf C., Paquette D., Pugsley M.K., Ratcliffe S., Redfern W.S., Stevens J., et al. Safety Pharmacology Investigations on the Nervous System: An Industry Survey. J. Pharmacol. Toxicol. Methods. 2016;81:37–46. doi: 10.1016/j.vascn.2016.06.001. - DOI - PubMed
-
- Kreir M., Van Deuren B., Versweyveld S., De Bondt A., Van den Wyngaert I., Van der Linde H., Lu H.R., Teuns G., Gallacher D.J. Do in Vitro Assays in Rat Primary Neurons Predict Drug-Induced Seizure Liability in Humans? Toxicol. Appl. Pharmacol. 2018;346:45–57. doi: 10.1016/j.taap.2018.03.028. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

