A Satellite-Free Centromere in Equus przewalskii Chromosome 10
- PMID: 36835543
- PMCID: PMC9961726
- DOI: 10.3390/ijms24044134
A Satellite-Free Centromere in Equus przewalskii Chromosome 10
Abstract
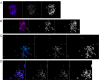
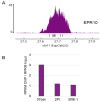
In mammals, centromeres are epigenetically specified by the histone H3 variant CENP-A and are typically associated with satellite DNA. We previously described the first example of a natural satellite-free centromere on Equus caballus chromosome 11 (ECA11) and, subsequently, on several chromosomes in other species of the genus Equus. We discovered that these satellite-free neocentromeres arose recently during evolution through centromere repositioning and/or chromosomal fusion, after inactivation of the ancestral centromere, where, in many cases, blocks of satellite sequences were maintained. Here, we investigated by FISH the chromosomal distribution of satellite DNA families in Equus przewalskii (EPR), demonstrating a good degree of conservation of the localization of the major horse satellite families 37cen and 2PI with the domestic horse. Moreover, we demonstrated, by ChIP-seq, that 37cen is the satellite bound by CENP-A and that the centromere of EPR10, the ortholog of ECA11, is devoid of satellite sequences. Our results confirm that these two species are closely related and that the event of centromere repositioning which gave rise to EPR10/ECA11 centromeres occurred in the common ancestor, before the separation of the two horse lineages.
Keywords: ChIP-seq; Equus przewalskii; centromere; karyotype evolution; satellite DNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
CENP-A and centromere evolution in equids.Chromosome Res. 2025 Jun 30;33(1):13. doi: 10.1007/s10577-025-09773-3. Chromosome Res. 2025. PMID: 40586953 Free PMC article. Review.
-
The localization of centromere protein A is conserved among tissues.Commun Biol. 2023 Sep 21;6(1):963. doi: 10.1038/s42003-023-05335-7. Commun Biol. 2023. PMID: 37735603 Free PMC article.
-
Robertsonian Fusion and Centromere Repositioning Contributed to the Formation of Satellite-free Centromeres During the Evolution of Zebras.Mol Biol Evol. 2022 Aug 3;39(8):msac162. doi: 10.1093/molbev/msac162. Mol Biol Evol. 2022. PMID: 35881460 Free PMC article.
-
Molecular Dynamics and Evolution of Centromeres in the Genus Equus.Int J Mol Sci. 2022 Apr 10;23(8):4183. doi: 10.3390/ijms23084183. Int J Mol Sci. 2022. PMID: 35457002 Free PMC article. Review.
-
The major horse satellite DNA family is associated with centromere competence.Mol Cytogenet. 2016 Apr 27;9:35. doi: 10.1186/s13039-016-0242-z. eCollection 2016. Mol Cytogenet. 2016. PMID: 27123044 Free PMC article.
Cited by
-
Transcription of a centromere-enriched retroelement and local retention of its RNA are significant features of the CENP-A chromatin landscape.Genome Biol. 2024 Nov 18;25(1):295. doi: 10.1186/s13059-024-03433-1. Genome Biol. 2024. PMID: 39558354 Free PMC article.
-
CENP-A and centromere evolution in equids.Chromosome Res. 2025 Jun 30;33(1):13. doi: 10.1007/s10577-025-09773-3. Chromosome Res. 2025. PMID: 40586953 Free PMC article. Review.
-
Mechanisms of Rapid Karyotype Evolution in Mammals.Genes (Basel). 2023 Dec 31;15(1):62. doi: 10.3390/genes15010062. Genes (Basel). 2023. PMID: 38254952 Free PMC article. Review.
-
The localization of centromere protein A is conserved among tissues.Commun Biol. 2023 Sep 21;6(1):963. doi: 10.1038/s42003-023-05335-7. Commun Biol. 2023. PMID: 37735603 Free PMC article.
-
CENP-A/CENP-B uncoupling in the evolutionary reshuffling of centromeres in equids.Genome Biol. 2025 Feb 6;26(1):23. doi: 10.1186/s13059-025-03490-0. Genome Biol. 2025. PMID: 39915813 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

