Small RNA and Degradome Sequencing in Floral Bud Reveal Roles of miRNAs in Dormancy Release of Chimonanthus praecox
- PMID: 36835618
- PMCID: PMC9964840
- DOI: 10.3390/ijms24044210
Small RNA and Degradome Sequencing in Floral Bud Reveal Roles of miRNAs in Dormancy Release of Chimonanthus praecox
Abstract
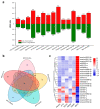
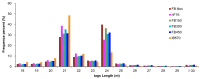
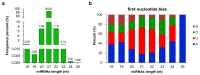
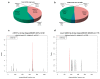
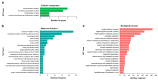
Chimonanthus praecox (wintersweet) is highly valued ornamentally and economically. Floral bud dormancy is an important biological characteristic in the life cycle of wintersweet, and a certain period of chilling accumulation is necessary for breaking floral bud dormancy. Understanding the mechanism of floral bud dormancy release is essential for developing measures against the effects of global warming. miRNAs play important roles in low-temperature regulation of flower bud dormancy through mechanisms that are unclear. In this study, small RNA and degradome sequencing were performed for wintersweet floral buds in dormancy and break stages for the first time. Small RNA sequencing identified 862 known and 402 novel miRNAs; 23 differentially expressed miRNAs (10 known and 13 novel) were screened via comparative analysis of breaking and other dormant floral bud samples. Degradome sequencing identified 1707 target genes of 21 differentially expressed miRNAs. The annotations of the predicted target genes showed that these miRNAs were mainly involved in the regulation of phytohormone metabolism and signal transduction, epigenetic modification, transcription factors, amino acid metabolism, and stress response, etc., during the dormancy release of wintersweet floral buds. These data provide an important foundation for further research on the mechanism of floral bud dormancy in wintersweet.
Keywords: Chimonanthus praecox; floral bud dormancy; high-throughput sequencing technology; miRNAs; target identification.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Transcriptomic analysis of flower development in wintersweet (Chimonanthus praecox).PLoS One. 2014 Jan 29;9(1):e86976. doi: 10.1371/journal.pone.0086976. eCollection 2014. PLoS One. 2014. PMID: 24489818 Free PMC article.
-
Small RNA and PARE sequencing in flower bud reveal the involvement of sRNAs in endodormancy release of Japanese pear (Pyrus pyrifolia 'Kosui').BMC Genomics. 2016 Mar 15;17:230. doi: 10.1186/s12864-016-2514-8. BMC Genomics. 2016. PMID: 26976036 Free PMC article.
-
RNA-Seq-based transcriptome analysis of dormant flower buds of Chinese cherry (Prunus pseudocerasus).Gene. 2015 Jan 25;555(2):362-76. doi: 10.1016/j.gene.2014.11.032. Epub 2014 Nov 15. Gene. 2015. PMID: 25447903
-
Molecular advances in bud dormancy in trees.J Exp Bot. 2024 Oct 16;75(19):6063-6075. doi: 10.1093/jxb/erae183. J Exp Bot. 2024. PMID: 38650362 Free PMC article. Review.
-
Interaction of Phytohormones and External Environmental Factors in the Regulation of the Bud Dormancy in Woody Plants.Int J Mol Sci. 2023 Dec 6;24(24):17200. doi: 10.3390/ijms242417200. Int J Mol Sci. 2023. PMID: 38139028 Free PMC article. Review.
Cited by
-
Small RNA-Seq to Unveil the miRNA Expression Patterns and Identify the Target Genes in Panax ginseng.Plants (Basel). 2023 Aug 27;12(17):3070. doi: 10.3390/plants12173070. Plants (Basel). 2023. PMID: 37687317 Free PMC article.
-
Unveiling the aesthetic secrets: exploring connections between genetic makeup, chemical, and environmental factors for enhancing/improving the color and fragrance/aroma of Chimonanthus praecox.PeerJ. 2024 Apr 19;12:e17238. doi: 10.7717/peerj.17238. eCollection 2024. PeerJ. 2024. PMID: 38650650 Free PMC article. Review.
-
Molecular Mechanisms of Seasonal Gene Expression in Trees.Int J Mol Sci. 2024 Jan 30;25(3):1666. doi: 10.3390/ijms25031666. Int J Mol Sci. 2024. PMID: 38338945 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

