Linking Lichen Metabolites to Genes: Emerging Concepts and Lessons from Molecular Biology and Metagenomics
- PMID: 36836275
- PMCID: PMC9964704
- DOI: 10.3390/jof9020160
Linking Lichen Metabolites to Genes: Emerging Concepts and Lessons from Molecular Biology and Metagenomics
Abstract
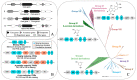
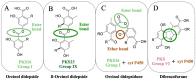
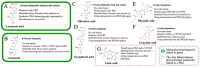
Lichen secondary metabolites have tremendous pharmaceutical and industrial potential. Although more than 1000 metabolites have been reported from lichens, less than 10 have been linked to the genes coding them. The current biosynthetic research focuses strongly on linking molecules to genes as this is fundamental to adapting the molecule for industrial application. Metagenomic-based gene discovery, which bypasses the challenges associated with culturing an organism, is a promising way forward to link secondary metabolites to genes in non-model, difficult-to-culture organisms. This approach is based on the amalgamation of the knowledge of the evolutionary relationships of the biosynthetic genes, the structure of the target molecule, and the biosynthetic machinery required for its synthesis. So far, metagenomic-based gene discovery is the predominant approach by which lichen metabolites have been linked to their genes. Although the structures of most of the lichen secondary metabolites are well-documented, a comprehensive review of the metabolites linked to their genes, strategies implemented to establish this link, and crucial takeaways from these studies is not available. In this review, I address the following knowledge gaps and, additionally, provide critical insights into the results of these studies, elaborating on the direct and serendipitous lessons that we have learned from them.
Keywords: atranorin; biosynthetic gene clusters; grayanic acid; gyrophoric acid; lecanoric acid; lichen metabolites; physodic/olivetoric acid; secondary metabolites; usnic acid.
Conflict of interest statement
The author declares no conflict of interest.
Figures
Similar articles
-
A Review of Anti-Cancer and Related Properties of Lichen-Extracts and Metabolites.Anticancer Agents Med Chem. 2022;22(1):115-142. doi: 10.2174/1871520621666210322094647. Anticancer Agents Med Chem. 2022. PMID: 34225637 Review.
-
Biological Effects of Gyrophoric Acid and Other Lichen Derived Metabolites, on Cell Proliferation, Apoptosis and Cell Signaling pathways.Chem Biol Interact. 2022 Jan 5;351:109768. doi: 10.1016/j.cbi.2021.109768. Epub 2021 Dec 3. Chem Biol Interact. 2022. PMID: 34864007 Free PMC article. Review.
-
Linking a Gene Cluster to Atranorin, a Major Cortical Substance of Lichens, through Genetic Dereplication and Heterologous Expression.mBio. 2021 Jun 29;12(3):e0111121. doi: 10.1128/mBio.01111-21. Epub 2021 Jun 22. mBio. 2021. PMID: 34154413 Free PMC article.
-
Larvicidal activity of some secondary lichen metabolites against the mosquito Culiseta longiareolata Macquart (Diptera: Culicidae).Nat Prod Res. 2012;26(4):350-5. doi: 10.1080/14786411003774296. Epub 2011 Jul 7. Nat Prod Res. 2012. PMID: 21452097
-
Lichen secondary metabolites affect growth of Physcomitrella patens by allelopathy.Protoplasma. 2017 May;254(3):1307-1315. doi: 10.1007/s00709-016-1022-7. Epub 2016 Sep 19. Protoplasma. 2017. PMID: 27645140
Cited by
-
Biosynthetic Potential of Hypogymnia Holobionts: Insights into Secondary Metabolite Pathways.J Fungi (Basel). 2023 May 9;9(5):546. doi: 10.3390/jof9050546. J Fungi (Basel). 2023. PMID: 37233257 Free PMC article.
-
Fungal BGCs for Production of Secondary Metabolites: Main Types, Central Roles in Strain Improvement, and Regulation According to the Piano Principle.Int J Mol Sci. 2023 Jul 6;24(13):11184. doi: 10.3390/ijms241311184. Int J Mol Sci. 2023. PMID: 37446362 Free PMC article. Review.
-
Genome-Wide Analysis of the Cytochrome P450 Monooxygenases in the Lichenized Fungi of the Class Lecanoromycetes.Microorganisms. 2023 Oct 19;11(10):2590. doi: 10.3390/microorganisms11102590. Microorganisms. 2023. PMID: 37894248 Free PMC article.
-
Lichens are a treasure chest of bioactive compounds: fact or fake?New Phytol. 2025 Apr;246(2):389-395. doi: 10.1111/nph.70034. Epub 2025 Feb 27. New Phytol. 2025. PMID: 40013383 Free PMC article. No abstract available.
-
Breaking into nature's secret medicine cabinet: lichens - a biochemical goldmine ready for discovery.New Phytol. 2025 Apr;246(2):437-449. doi: 10.1111/nph.70003. Epub 2025 Feb 26. New Phytol. 2025. PMID: 40007421 Free PMC article. Review.
References
-
- Boustie J., Grube M. Lichens—A Promising Source of Bioactive Secondary Metabolites. Plant Genet. Resour. 2005;3:273–287. doi: 10.1079/PGR200572. - DOI
Publication types
LinkOut - more resources
Full Text Sources

