Integrating Metabolomics and Gene Expression Underlying Potential Biomarkers Compounds Associated with Antioxidant Activity in Southern Grape Seeds
- PMID: 36837828
- PMCID: PMC9963462
- DOI: 10.3390/metabo13020210
Integrating Metabolomics and Gene Expression Underlying Potential Biomarkers Compounds Associated with Antioxidant Activity in Southern Grape Seeds
Abstract
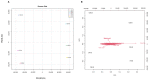
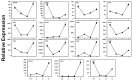
Different southern grape (Muscadine) genotypes (Muscadinia rotundifolia Michx.) were evaluated for their contents of metabolites in ripe berries. The metabolome study identified 331 metabolites in ripening skin and seed tissues. The major chemical groups were organic acids, fatty acyls, polyketides, and organic heterocycle compounds. The metabolic pathways of the identified metabolite were mainly arginine biosynthesis, D-glutamine, D-glutamate metabolism, alanine, aspartate metabolism, aminoacyl-tRNA biosynthesis, and citrate cycle. Principal component analysis indicated that catechin, gallic acid, and epicatechin-3-gallate were the main metabolites existing in muscadine seed extracts. However, citramalic and malic acids were the main metabolites contributing to muscadine skin extracts. Partial least-squares discriminant analysis (VIP > 1) described 25 key compounds indicating the metabolome in muscadine tissues (skin and seed). Correlation analysis among the 25 compounds and oxidation inhibition activities identified five biomarker compounds that were associated with antioxidant activity. Catechin, gallic acid, epicatechin-3-gallate, fertaric acid, and procyanidin B1 were highly associated with DPPH, FRAP, CUPRAC, and ABTS. The five biomarker compounds were significantly accumulated in the seed relative to the skin tissues. An evaluation of 15 antioxidant-related genes represented by the 3-dehydroquinate dehydratase (DHD), shikimate kinase (SK), chalcone synthase (CHS), anthocyanidin reductase (ANR), laccase (LAC), phenylalanine ammonia-lyase (PAL), dihydroflavonol 4-reductase (DFR), 3-dehydroquinate synthase (DHQS), chorismate mutase (CM), flavanone-3-hydroxylase (F3H), cinnamoyl-CoA reductase (CCR), cinnamyl alcohol dehydrogenase (CAD), leucoanthocyanidin reductase (LAR), gallate 1-β-glucosyltransferase (UGT), and anthocyanidin 3-O-glucosyltransferase (UFGT) encode critical enzymes related to polyphenolics pathway throughout four developmental stages (fruit-set FS, véraison V, ripe-skin R, and ripe-seed; S) in the C5 genotype demonstrated the dramatic accumulation of all transcripts in seed tissue or a developmental stage-dependent manner. Our findings suggested that muscadine grape seeds contain essential metabolites that could attract the attention of those interested in the pharmaceutical sector and the plant breeders to develop new varieties with high nutraceutical value.
Keywords: antioxidant; biomarker compounds; genes; metabolome; muscadine genotypes; seed.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- Jayaprakasha G.K., Singh R.P., Sakariah K.K. Antioxidant activity of grape seed (Vitis vinifera) extracts on peroxidation models in vitro. Food Chem. 2021;73:285–290. doi: 10.1016/S0308-8146(00)00298-3. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

