Severe Acute Respiratory Syndrome Coronavirus 2 Receptor (Human Angiotensin-Converting Enzyme 2) Binding Inhibition Assay: A Rapid, High-Throughput Assay Useful for Vaccine Immunogenicity Evaluation
- PMID: 36838333
- PMCID: PMC9965183
- DOI: 10.3390/microorganisms11020368
Severe Acute Respiratory Syndrome Coronavirus 2 Receptor (Human Angiotensin-Converting Enzyme 2) Binding Inhibition Assay: A Rapid, High-Throughput Assay Useful for Vaccine Immunogenicity Evaluation
Abstract
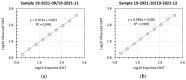
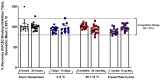
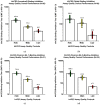
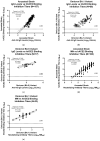
Emerging variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) show immune evasion of vaccine-derived immunity, highlighting the need for better clinical immunogenicity biomarkers. To address this need, an enzyme-linked immunosorbent assay-based, human angiotensin-converting enzyme 2 (hACE2) binding inhibition assay was developed to measure antibodies against the ancestral strain of SARS-CoV-2 and was validated for precision, specificity, linearity, and other parameters. This assay measures the inhibition of SARS-CoV-2 spike (S) protein binding to the receptor, hACE2, by serum from vaccine clinical trials. Inter- and intra-assay precision, specificity, linearity, lower limit of quantitation, and assay robustness parameters successfully met the acceptance criteria. Heme and lipid matrix effects showed minimal interference on the assay. Samples were stable for testing in the assay even with 8 freeze/thaws and up to 24 months in -80 °C storage. The assay was also adapted for variants (Delta and Omicron BA.1/BA.5), with similar validation results. The hACE2 assay showed significant correlation with anti-recombinant S immunoglobulin G levels and neutralizing antibody titers. This assay provides a rapid, high-throughput option to evaluate vaccine immunogenicity. Along with other clinical biomarkers, it can provide valuable insights into immune evasion and correlates of protection and enable vaccine development against emerging COVID-19 variants.
Keywords: COVID-19; SARS-CoV-2; assay validation; correlate of protection; human angiotensin-converting enzyme 2 (hACE2); immune evasion; immunogenicity; neutralizing antibody titers.
Conflict of interest statement
All the authors are employees and stockholders of Novavax, Inc.
Figures

References
-
- Centers for Disease Control and Prevention David J. Spencer CDC Museum—COVID-19 Timeline. [(accessed on 24 August 2022)]; Available online: https://www.cdc.gov/museum/timeline/covid19.html.
-
- Fiolet T., Kherabi Y., MacDonald C.J., Ghosn J., Peiffer-Smadja N. Comparing COVID-19 vaccines for their characteristics, efficacy and effectiveness against SARS-CoV-2 and variants of concern: A narrative review. Clin. Microbiol. Infect. 2022;28:202–221. doi: 10.1016/j.cmi.2021.10.005. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

