Active Sulfate-Reducing Bacterial Community in the Camel Gut
- PMID: 36838366
- PMCID: PMC9963290
- DOI: 10.3390/microorganisms11020401
Active Sulfate-Reducing Bacterial Community in the Camel Gut
Abstract
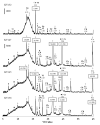
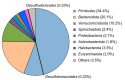
The diversity and activity of sulfate-reducing bacteria (SRB) in the camel gut remains largely unexplored. An abundant SRB community has been previously revealed in the feces of Bactrian camels (Camelus bactrianus). This study aims to combine the 16S rRNA gene profiling, sulfate reduction rate (SRR) measurement with a radioactive tracer, and targeted cultivation to shed light on SRB activity in the camel gut. Fresh feces of 55 domestic Bactrian camels grazing freely on semi-arid mountain pastures in the Kosh-Agach district of the Russian Altai area were analyzed. Feces were sampled in early winter at an ambient temperature of -15 °C, which prevented possible contamination. SRR values measured with a radioactive tracer in feces were relatively high and ranged from 0.018 to 0.168 nmol S cm-3 day-1. The 16S rRNA gene profiles revealed the presence of Gram-negative Desulfovibrionaceae and spore-forming Desulfotomaculaceae. Targeted isolation allowed us to obtain four pure culture isolates belonging to Desulfovibrio and Desulforamulus. An active SRB community may affect the iron and copper availability in the camel intestine due to metal ions precipitation in the form of sparingly soluble sulfides. The copper-iron sulfide, chalcopyrite (CuFeS2), was detected by X-ray diffraction in 36 out of 55 analyzed camel feces. In semi-arid areas, gypsum, like other evaporite sulfates, can be used as a solid-phase electron acceptor for sulfate reduction in the camel gastrointestinal tract.
Keywords: Desulforamulus; Desulfovibrio; biogenic chalcopyrite; camels; gut microbiota; sulfate-reduction.
Conflict of interest statement
The authors declare that there are no conflict of interest.
Figures




References
-
- Deplancke B., Hristova K.R., Oakley H.A., McCracken V.J., Aminov R., Mackie R.I., Gaskins H.R. Molecular Ecological Analysis of the Succession and Diversity of Sulfate-Reducing Bacteria in the Mouse Gastrointestinal Tract. Appl. Environ. Microbiol. 2000;66:2166–2174. doi: 10.1128/AEM.66.5.2166-2174.2000. - DOI - PMC - PubMed
-
- Macfarlane G.T., Cummings J.H., Macfarlane S. Sulphate-reducing bacteria and the human large intestine. In: Barton L.L., Hamilton W.A., editors. Sulphate-Reducing Bacteria. Environmental and Engineered Systems. Cambridge University Press; Cambridge, UK: 2007. pp. 503–523.
Grants and funding
LinkOut - more resources
Full Text Sources

