Protoplast Preparation for Algal Single-Cell Omics Sequencing
- PMID: 36838504
- PMCID: PMC9962006
- DOI: 10.3390/microorganisms11020538
Protoplast Preparation for Algal Single-Cell Omics Sequencing
Abstract
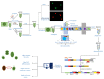
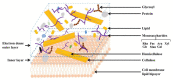
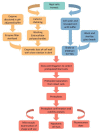
Single-cell sequencing (SCS) is an evolutionary technique for conducting life science research, providing the highest genome-sale throughput and single-cell resolution and unprecedented capabilities in addressing mechanistic and operational questions. Unfortunately, the current SCS pipeline cannot be directly applied to algal research as algal cells have cell walls, which makes RNA extraction hard for the current SCS platforms. Fortunately, effective methods are available for producing algal protoplasts (cells without cell walls), which can be directly fed into current SCS pipelines. In this review, we first summarize the cell wall structure and chemical composition of algal cell walls, particularly in Chlorophyta, then summarize the advances made in preparing algal protoplasts using physical, chemical, and biological methods, followed by specific cases of algal protoplast production in some commonly used eukaryotic algae. This review provides a timely primer to those interested in applying SCS in eukaryotic algal research.
Keywords: Chlorella vulgaris; algal protoplast; bead; cell wall; cellulose; enzymatic lysis; sequencing; single-cell.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Experimental evidence for enzymatic cell wall dissolution in a microbial protoplast feeder (Orciraptor agilis, Viridiraptoridae).BMC Biol. 2022 Dec 5;20(1):267. doi: 10.1186/s12915-022-01478-x. BMC Biol. 2022. PMID: 36464670 Free PMC article.
-
Protoplast preparation from enriched flagellates and resting cells of Haematococcus pluvialis.J Appl Microbiol. 2018 Feb;124(2):469-479. doi: 10.1111/jam.13643. J Appl Microbiol. 2018. PMID: 29154453
-
Insights into cell wall disintegration of Chlorella vulgaris.PLoS One. 2022 Jan 14;17(1):e0262500. doi: 10.1371/journal.pone.0262500. eCollection 2022. PLoS One. 2022. PMID: 35030225 Free PMC article.
-
Viruses of Eukaryotic Algae: Diversity, Methods for Detection, and Future Directions.Viruses. 2018 Sep 11;10(9):487. doi: 10.3390/v10090487. Viruses. 2018. PMID: 30208617 Free PMC article. Review.
-
Chemical Synthesis of Oligosaccharides Related to the Cell Walls of Plants and Algae.Chem Rev. 2017 Sep 13;117(17):11337-11405. doi: 10.1021/acs.chemrev.7b00162. Epub 2017 Aug 9. Chem Rev. 2017. PMID: 28792736 Review.
Cited by
-
An Innovative Bioremediation Approach to Heavy Metal Removal: Combined Application of Chlorella vulgaris and Amine-Functionalized MgFe2O4 Nanoparticles in Industrial Wastewater Treatment.Int J Mol Sci. 2025 Jun 7;26(12):5467. doi: 10.3390/ijms26125467. Int J Mol Sci. 2025. PMID: 40564931 Free PMC article.
-
Applications of single‑cell omics and spatial transcriptomics technologies in gastric cancer (Review).Oncol Lett. 2024 Feb 14;27(4):152. doi: 10.3892/ol.2024.14285. eCollection 2024 Apr. Oncol Lett. 2024. PMID: 38406595 Free PMC article. Review.
References
-
- Maynard K.R., Collado-Torres L., Weber L.M., Uytingco C., Barry B.K., Williams S.R., Catallini J.L., Tran M.N., Besich Z., Tippani M., et al. Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex. Nat. Neurosci. 2021;24:425–436. doi: 10.1038/s41593-020-00787-0. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

