Bio-Electroanalysis Performance of Heme Redox-Center for π- π Interaction Bonding of a Methylene Blue-Graphene Modified Electrode
- PMID: 36839114
- PMCID: PMC9963319
- DOI: 10.3390/nano13040745
Bio-Electroanalysis Performance of Heme Redox-Center for π- π Interaction Bonding of a Methylene Blue-Graphene Modified Electrode
Abstract
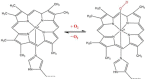
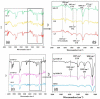
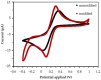
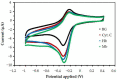
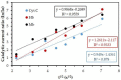
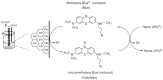
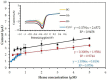
Hemeprotein detection has motivated extensive research on the direct reaction of a heme molecule and a redox dye. The present study used methylene blue as both donor and acceptor for a redox reaction. First, the solid phases of methylene blue (MB) and graphene (GP) formed a π-π interaction bond at the aromatic rings. The conductivity of GP was better than that of carbon in a carbon electrode (CE). Then, the working CE was modified using strong adsorption of MB/GP on the electrode surface. The surface of the electrode was investigated using a modified and an unmodified electrode. The electrode's properties were studied using voltammograms of redox couple K3[Fe(CN)6]3-/4-. Its reaction was used to find the active area of the modified electrode, which was 1.76 times bigger than that of the unmodified electrode. The surface coverage values of the modified and unmodified electrodes were 8.17 × 10-6 and 1.53 × 10-5 mol/cm2, respectively. This research also studied the application of hemeprotein detection. Hemoglobin (Hb), myoglobin (Mb), and cytochrome c (Cyt. C) were studied by the reaction of Fe (III/II) at the heme-redox center. The electrocatalytic reaction between MB/GP and hemeproteins produced an anodic peak at 0.35 V for Hb, Mb, and Cyt. C. This nanohybrid film enhanced electron transfer between protein molecules and the modified carbon electrode. The amperometric measurements show that the limit of detection was 0.2 µM, 0.3 µM, and 0.1 µM for Hb, Mb, and Cyt. C, respectively. The measurement spanned a linear range of 0.2 µM to 5 µM, 0.3 µM to 5 µM, and 0.1 µM to 0.7 µM for Hb, Mb, and Cyt. C, respectively. Hb showed the lowest sensitivity compared with Mb and Cyt. C due to the role of steric hindrance in the hemeprotein specificity structure. This study offers a simple and efficient fabrication platform for electrochemical sensors for hemeproteins. When compared to other complex immobilization processes, the fabrication method for this sensor has many benefits, including no need for special chemicals and easy preparation and electrode modification-both of which are crucial for the development of electrochemical sensing devices.
Keywords: electrochemical sensor; graphene; heme; methylene blue; modified electrode.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Electrochemical behavior of biocatalytical composite based on heme-proteins, didodecyldimethylammonium bromide and room-temperature ionic liquid.Anal Chim Acta. 2010 Mar 17;663(1):19-26. doi: 10.1016/j.aca.2010.01.015. Epub 2010 Jan 18. Anal Chim Acta. 2010. PMID: 20172091
-
Immobilization of hemoglobin on electrodeposited cobalt-oxide nanoparticles: direct voltammetry and electrocatalytic activity.Biophys Chem. 2007 Nov;130(3):122-31. doi: 10.1016/j.bpc.2007.08.004. Epub 2007 Aug 24. Biophys Chem. 2007. PMID: 17825977
-
Direct electrochemistry of hemoglobin and myoglobin at didodecyldimethylammonium bromide-modified powder microelectrode and application for electrochemical detection of nitric oxide.Anal Chim Acta. 2008 Jan 21;607(1):30-6. doi: 10.1016/j.aca.2007.11.038. Epub 2007 Nov 28. Anal Chim Acta. 2008. PMID: 18155406
-
Mixed hemi/ad-micelles coated magnetic nanoparticles for the entrapment of hemoglobin at the surface of a screen-printed carbon electrode and its direct electrochemistry and electrocatalysis.Biosens Bioelectron. 2015 Dec 15;74:518-25. doi: 10.1016/j.bios.2015.07.001. Epub 2015 Jul 5. Biosens Bioelectron. 2015. PMID: 26183073
-
Hemoglobin and Myoglobin as Reducing Agents in Biological Systems. Redox Reactions of Globins with Copper and Iron Salts and Complexes.Biochemistry (Mosc). 2016 Dec;81(13):1735-1753. doi: 10.1134/S0006297916130101. Biochemistry (Mosc). 2016. PMID: 28260494 Review.
References
-
- Lodish H., Berk A., Kaiser C.-A., Krieger M., Scott M.-P., Bretscher A., Matsudaira P. Molecular Cell Biology. 6th ed. W.H. Freeman and Company; New York, NY, USA: 2008.
-
- Nelson D.-L., Cox M.-M. Principle of Biochemistry. W.H. Freeman and Company; New York, NY, USA: 2008.
-
- Pough F.-H., Janis C.-M., Heiser J.-B. Vertebrate Life Boston. Pearson Benjamin Cummings; San Francisco, CA, USA: 2009.
-
- Lei C., Woollenberger U., Bistolas N., Guiseppi-Elis A., Scheller F.W. Analytical Bioanalytical. Chemistry. 2002;372:235–239. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

