The DarT/DarG Toxin-Antitoxin ADP-Ribosylation System as a Novel Target for a Rational Design of Innovative Antimicrobial Strategies
- PMID: 36839512
- PMCID: PMC9967889
- DOI: 10.3390/pathogens12020240
The DarT/DarG Toxin-Antitoxin ADP-Ribosylation System as a Novel Target for a Rational Design of Innovative Antimicrobial Strategies
Abstract
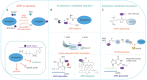
The chemical modification of cellular macromolecules by the transfer of ADP-ribose unit(s), known as ADP-ribosylation, is an ancient homeostatic and stress response control system. Highly conserved across the evolution, ADP-ribosyltransferases and ADP-ribosylhydrolases control ADP-ribosylation signalling and cellular responses. In addition to proteins, both prokaryotic and eukaryotic transferases can covalently link ADP-ribosylation to different conformations of nucleic acids, thus highlighting the evolutionary conservation of archaic stress response mechanisms. Here, we report several structural and functional aspects of DNA ADP-ribosylation modification controlled by the prototype DarT and DarG pair, which show ADP-ribosyltransferase and hydrolase activity, respectively. DarT/DarG is a toxin-antitoxin system conserved in many bacterial pathogens, for example in Mycobacterium tuberculosis, which regulates two clinically important processes for human health, namely, growth control and the anti-phage response. The chemical modulation of the DarT/DarG system by selective inhibitors may thus represent an exciting strategy to tackle resistance to current antimicrobial therapies.
Keywords: ADP-ribosylation; DNA modification; DarT/DarG; antimicrobial resistance; cell growth; phage defence; toxin–antitoxin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Streptomyces coelicolor macrodomain hydrolase SCO6735 cleaves thymidine-linked ADP-ribosylation of DNA.Comput Struct Biotechnol J. 2022 Aug 8;20:4337-4350. doi: 10.1016/j.csbj.2022.08.002. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36051881 Free PMC article.
-
Molecular basis for DarT ADP-ribosylation of a DNA base.Nature. 2021 Aug;596(7873):597-602. doi: 10.1038/s41586-021-03825-4. Epub 2021 Aug 18. Nature. 2021. PMID: 34408320
-
Structural insights into DarT toxin neutralization by cognate DarG antitoxin: ssDNA mimicry by DarG C-terminal domain keeps the DarT toxin inhibited.Structure. 2023 Jul 6;31(7):780-789.e4. doi: 10.1016/j.str.2023.04.008. Epub 2023 May 10. Structure. 2023. PMID: 37167974
-
ADP-ribosylation of RNA and DNA: from in vitro characterization to in vivo function.Nucleic Acids Res. 2021 Apr 19;49(7):3634-3650. doi: 10.1093/nar/gkab136. Nucleic Acids Res. 2021. PMID: 33693930 Free PMC article. Review.
-
A role of intracellular mono-ADP-ribosylation in cancer biology.FEBS J. 2013 Aug;280(15):3551-62. doi: 10.1111/febs.12290. Epub 2013 May 10. FEBS J. 2013. PMID: 23590234 Review.
Cited by
-
Nucleotidyltransferase toxin MenT extends aminoacyl acceptor ends of serine tRNAs to control Mycobacterium tuberculosis growth.Nat Commun. 2024 Nov 6;15(1):9596. doi: 10.1038/s41467-024-53931-w. Nat Commun. 2024. PMID: 39505885 Free PMC article.
-
Targeting the Ubiquitin-Proteasome System and Recent Advances in Cancer Therapy.Cells. 2023 Dec 22;13(1):29. doi: 10.3390/cells13010029. Cells. 2023. PMID: 38201233 Free PMC article. Review.
-
MenT nucleotidyltransferase toxins extend tRNA acceptor stems and can be inhibited by asymmetrical antitoxin binding.Nat Commun. 2023 Aug 17;14(1):4644. doi: 10.1038/s41467-023-40264-3. Nat Commun. 2023. PMID: 37591829 Free PMC article.
-
Discovery of reversing enzymes for RNA ADP-ribosylation reveals a possible defence module against toxic attack.Nucleic Acids Res. 2025 Feb 8;53(4):gkaf069. doi: 10.1093/nar/gkaf069. Nucleic Acids Res. 2025. PMID: 39964479 Free PMC article.
-
Identification and characterization of a novel type II toxin-antitoxin system in Aeromonas veronii.Arch Microbiol. 2024 Aug 17;206(9):381. doi: 10.1007/s00203-024-04101-5. Arch Microbiol. 2024. PMID: 39153128
References
Grants and funding
LinkOut - more resources
Full Text Sources

