Antiviral Mechanism of Virucidal Sialic Acid Modified Cyclodextrin
- PMID: 36839904
- PMCID: PMC9965221
- DOI: 10.3390/pharmaceutics15020582
Antiviral Mechanism of Virucidal Sialic Acid Modified Cyclodextrin
Abstract
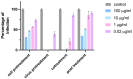
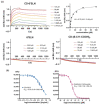
We have reported that CD-6'SLN [6-sialyllactosamine (6'SLN)-modified β-cyclodextrin (CD)] can be a potential anti-influenza drug because it irreversibly deactivates virions. Indeed, in vivo, CD-6'SLN improved mice survival in an H1N1 infection model even when administered 24 h post-infection. Although CD-6'SLN was designed to target the viral envelope protein hemagglutinin (HA), a natural receptor of 6'SLN, it remains unclear whether other targets exist. In this study, we confirm that CD-6'SLN inhibits the influenza virus through an extracellular mechanism by interacting with HA, but not with neuraminidase (NA), despite the latter also having a binding pocket for the sialyl group. We find that CD-6'SLN interacts with the viral envelope as it elicits the release of a fluorophore embedded in the membrane. Two similar compounds were designed to test separately the effect of 6'SLN and of the undecyl moiety that links the CD to 6'SLN. Neither showed any interaction with the membrane nor the irreversible viral inhibition (virucidal), confirming that both components are essential to membrane interaction and virucidal action. Unlike similar antiviral cyclodextrins developed against other viruses, CD-6'SLN was not able to decapsulate viral RNA. Our findings support that combining viral protein-specific epitopes with hydrophobic linkers provides a strategy for developing antiviral drugs with a virucidal mechanism.
Keywords: 6′SLN; antiviral mechanism; hemagglutinin; hydrophobic linker; influenza; membrane interaction; virucidal.
Conflict of interest statement
F.S. and P.H.J.S. are co-founders of Asterivir, a start-up company that is trying to develop novel antivirals. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Figures



References
-
- Iuliano A.D., Roguski K.M., Chang H.H., Muscatello D.J., Palekar R., Tempia S., Cohen C., Gran J.M., Schanzer D., Cowling B.J., et al. Estimates of global seasonal influenza-associated respiratory mortality: A modelling study. Lancet. 2018;391:1285–1300. doi: 10.1016/S0140-6736(17)33293-2. - DOI - PMC - PubMed
-
- Omoto S., Speranzini V., Hashimoto T., Noshi T., Yamaguchi H., Kawai M., Kawaguchi K., Uehara T., Shishido T., Naito A., et al. Characterization of influenza virus variants induced by treatment with the endonuclease inhibitor baloxavir marboxil. Sci. Rep. 2018;8:9633. doi: 10.1038/s41598-018-27890-4. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

