Drugging Hijacked Kinase Pathways in Pediatric Oncology: Opportunities and Current Scenario
- PMID: 36839989
- PMCID: PMC9966033
- DOI: 10.3390/pharmaceutics15020664
Drugging Hijacked Kinase Pathways in Pediatric Oncology: Opportunities and Current Scenario
Abstract
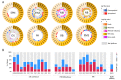
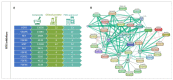
Childhood cancer is considered rare, corresponding to ~3% of all malignant neoplasms in the human population. The World Health Organization (WHO) reports a universal occurrence of more than 15 cases per 100,000 inhabitants around the globe, and despite improvements in diagnosis, treatment and supportive care, one child dies of cancer every 3 min. Consequently, more efficient, selective and affordable therapeutics are still needed in order to improve outcomes and avoid long-term sequelae. Alterations in kinases' functionality is a trademark of cancer and the concept of exploiting them as drug targets has burgeoned in academia and in the pharmaceutical industry of the 21st century. Consequently, an increasing plethora of inhibitors has emerged. In the present study, the expression patterns of a selected group of kinases (including tyrosine receptors, members of the PI3K/AKT/mTOR and MAPK pathways, coordinators of cell cycle progression, and chromosome segregation) and their correlation with clinical outcomes in pediatric solid tumors were accessed through the R2: Genomics Analysis and Visualization Platform and by a thorough search of published literature. To further illustrate the importance of kinase dysregulation in the pathophysiology of pediatric cancer, we analyzed the vulnerability of different cancer cell lines against their inhibition through the Cancer Dependency Map portal, and performed a search for kinase-targeted compounds with approval and clinical applicability through the CanSAR knowledgebase. Finally, we provide a detailed literature review of a considerable set of small molecules that mitigate kinase activity under experimental testing and clinical trials for the treatment of pediatric tumors, while discuss critical challenges that must be overcome before translation into clinical options, including the absence of compounds designed specifically for childhood tumors which often show differential mutational burdens, intrinsic and acquired resistance, lack of selectivity and adverse effects on a growing organism.
Keywords: chemical inhibitors; childhood cancer; clinical trials; kinases.
Conflict of interest statement
The authors declare no conflict of interests.
Figures




References
-
- Instituto Nacional de Câncer (Brazil) Coordenação de Prevenção e Vigilância and Sociedade Brasileira de Oncologia Pediátrica, Câncer na Criança e no Adolescente no Brasil: Dados dos Registros de Base Populacional e de Mortalidade. Ministério da Saúde, Instituto Nacional de Câncer–INCA; Brasília, Brazil: 2008.
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

