Ononin alleviates DSS-induced colitis through inhibiting NLRP3 inflammasome via triggering mitophagy
- PMID: 36840499
- PMCID: PMC9910166
- DOI: 10.1002/iid3.776
Ononin alleviates DSS-induced colitis through inhibiting NLRP3 inflammasome via triggering mitophagy
Abstract
Background: Ononin, a flavonoid isolated from Astragalus membranaceus root, is the active ingredient of A. membranaceus and has potential anti-inflammatory properties, but its effect on colitis is unclear.
Aims: This study aimed to explore the anticolitis effect of Ononin by establishing a colitis model in mice induced by dextran sulfate sodium (DSS).
Methods: Male C57BL/6 mice were provided DSS, then treated with Ononin (10, 20, 40 mg/kg) or 5-ASA (40 mg/kg). The colitis symptoms were observed, the disease activity index (DAI) score were recorded daily, and colonic inflammation was evaluted by histopathological scoring. The expression of cytokines, inflammatory mediators, and mitophagy/NLRP3 inflammasome-related proteins were measured.
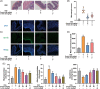
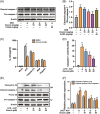
Results: Ononin significantly alleviated weight loss and colon shortening in mice with colitis (p < .01). Moreover, Ononin decreased the production of inflammatory cytokines and mediators associated with colitis (p < .05). In addition, Ononin inhibited macrophages infiltration and reduced caspase-1 activation in colitis mice. Caspase-1 activation is closely related to the NLRP3 inflammasome. Therefore, we investigated the effect of Ononin on NLRP3 inflammasome in vitro. The relevant results confirmed that Ononin inhibited NLRP3 inflammasome activation and inhibited mitochondrial damage (p < .05). Further studies revealed that Ononin inhibited mitochondrial damage through triggering mitophagy (p < .05).
Conclusion: Ononin alleviates DSS-induced colitis by activating mitophagy to inhibit NLRP3 inflammasome.
Keywords: NLRP3 inflammasome; Ononin; colitis; mitochondrial damage; mitophagy.
© 2023 The Authors. Immunity, Inflammation and Disease published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

