Effect of tRNA Maturase Depletion on Levels and Stabilities of Ribosome Assembly Cofactor and Other mRNAs in Bacillus subtilis
- PMID: 36840557
- PMCID: PMC10100781
- DOI: 10.1128/spectrum.05134-22
Effect of tRNA Maturase Depletion on Levels and Stabilities of Ribosome Assembly Cofactor and Other mRNAs in Bacillus subtilis
Abstract
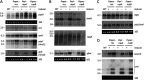
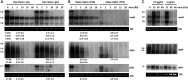
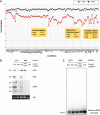
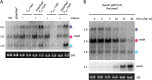
The impact of translation on mRNA stability can be varied, ranging from a protective effect of ribosomes that shield mRNA from RNases to preferentially exposing sites of RNase cleavage. These effects can change depending on whether ribosomes are actively moving along the mRNA or stalled at particular sequences or structures or awaiting charged tRNAs. We recently observed that depleting Bacillus subtilis cells of their tRNA maturation enzymes RNase P and RNase Z led to altered mRNA levels of a number of assembly factors involved in the biogenesis of the 30S ribosomal subunit. Here, we extended this study to other assembly factor and non-assembly factor mRNAs in B. subtilis. We additionally identified multiple transcriptional and translational layers of regulation of the rimM operon mRNA that occur in response to the depletion of functional tRNAs. IMPORTANCE The passage of ribosomes across individual mRNAs during translation can have different effects on their degradation, ranging from a protective effect by shielding from ribonucleases to, in some cases, making the mRNA more vulnerable to RNase action. We recently showed that some mRNAs coding for proteins involved in ribosome assembly were highly sensitive to the availability of functional tRNA. Using strains depleted of the major tRNA processing enzymes RNase P and RNase Z, we expanded this observation to a wider set of mRNAs, including some unrelated to ribosome biogenesis. We characterized the impact of tRNA maturase depletion on the rimM operon mRNA and show that it is highly complex, with multiple levels of transcriptional and posttranscriptional effects coming into play.
Keywords: (p)ppGpp; RNA degradation; chloramphenicol; tRNA processing; translation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Interaction of the Bacillus subtilis RNase P with the 30S ribosomal subunit.RNA. 2004 Mar;10(3):482-92. doi: 10.1261/rna.5163104. RNA. 2004. PMID: 14970393 Free PMC article.
-
Dynamic Membrane Localization of RNase Y in Bacillus subtilis.mBio. 2020 Feb 18;11(1):e03337-19. doi: 10.1128/mBio.03337-19. mBio. 2020. PMID: 32071272 Free PMC article.
-
Maturation of polycistronic mRNAs by the endoribonuclease RNase Y and its associated Y-complex in Bacillus subtilis.Proc Natl Acad Sci U S A. 2018 Jun 12;115(24):E5585-E5594. doi: 10.1073/pnas.1803283115. Epub 2018 May 24. Proc Natl Acad Sci U S A. 2018. PMID: 29794222 Free PMC article.
-
RNA processing and degradation in Bacillus subtilis.Microbiol Mol Biol Rev. 2003 Jun;67(2):157-74, table of contents. doi: 10.1128/MMBR.67.2.157-174.2003. Microbiol Mol Biol Rev. 2003. PMID: 12794188 Free PMC article. Review.
-
The critical role of RNA processing and degradation in the control of gene expression.FEMS Microbiol Rev. 2010 Sep;34(5):883-923. doi: 10.1111/j.1574-6976.2010.00242.x. Epub 2010 Jun 24. FEMS Microbiol Rev. 2010. PMID: 20659169 Review.
Cited by
-
Antibacterial activity and antibacterial mechanism of flavaspidic acid BB against Staphylococcus haemelyticus.BMC Microbiol. 2023 Sep 29;23(1):276. doi: 10.1186/s12866-023-02997-5. BMC Microbiol. 2023. PMID: 37773054 Free PMC article.
-
The DnaJK chaperone of Bacillus subtilis post-transcriptionally regulates gene expression through the YlxR(RnpM)/RNase P complex.mBio. 2025 Mar 12;16(3):e0405324. doi: 10.1128/mbio.04053-24. Epub 2025 Feb 11. mBio. 2025. PMID: 39932325 Free PMC article.
References
-
- Braun F, Condon C. 2019. RNA processing, p 164–177. In Schmidt TM (ed), Encyclopedia of microbiology, 4th ed. Elsevier, Amsterdam, The Netherlands.
LinkOut - more resources
Full Text Sources

