Structural insights into the binding of bS1 to the ribosome
- PMID: 36840711
- PMCID: PMC10123108
- DOI: 10.1093/nar/gkad126
Structural insights into the binding of bS1 to the ribosome
Abstract
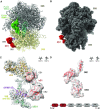
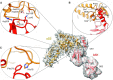
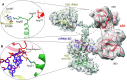
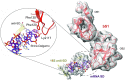
The multidomain ribosomal protein bS1 is the biggest and the most flexible and dynamic protein in the 30S small subunit. Despite being essential for mRNA recruitment and its primary role in the accommodation of the start codon within the decoding centre, there has not yet been a high-resolution description of its structure. Here, we present a 3D atomic model of OB1 and OB2, bS1's first two N-terminal domains, bound to an elongation-competent 70S ribosome. Our structure reveals that, as previously reported, bS1 is anchored both by a π-stacking to the 30S subunit and via a salt bridge with the Zn2+ pocket of bS1. These contacts are further stabilized by other interactions with additional residues on OB1. Our model also shows a new conformation of OB2, interacting with the Shine-Dalgarno portion of the mRNA. This study confirms that OB1 plays an anchoring role, but also highlights a novel function for OB2, which is directly involved in the modulation and support of mRNA binding and accommodation on the ribosome.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
Role of Ribosomal Protein bS1 in Orthogonal mRNA Start Codon Selection.Biochemistry. 2025 Feb 4;64(3):710-718. doi: 10.1021/acs.biochem.4c00688. Epub 2025 Jan 24. Biochemistry. 2025. PMID: 39854700 Free PMC article.
-
Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1.Nat Microbiol. 2018 Oct;3(10):1115-1121. doi: 10.1038/s41564-018-0237-0. Epub 2018 Sep 3. Nat Microbiol. 2018. PMID: 30177741
-
Molecular basis of mRNA delivery to the bacterial ribosome.Science. 2024 Nov 29;386(6725):eado8476. doi: 10.1126/science.ado8476. Epub 2024 Nov 29. Science. 2024. PMID: 39607923
-
Structural basis for messenger RNA movement on the ribosome.Nature. 2006 Nov 16;444(7117):391-4. doi: 10.1038/nature05281. Epub 2006 Oct 18. Nature. 2006. PMID: 17051149
-
Assembly of bacterial ribosomes.Annu Rev Biochem. 2011;80:501-26. doi: 10.1146/annurev-biochem-062608-160432. Annu Rev Biochem. 2011. PMID: 21529161 Review.
Cited by
-
Extraribosomal Functions of Bacterial Ribosomal Proteins-An Update, 2023.Int J Mol Sci. 2024 Mar 3;25(5):2957. doi: 10.3390/ijms25052957. Int J Mol Sci. 2024. PMID: 38474204 Free PMC article. Review.
-
Role of Ribosomal Protein bS1 in Orthogonal mRNA Start Codon Selection.Biochemistry. 2025 Feb 4;64(3):710-718. doi: 10.1021/acs.biochem.4c00688. Epub 2025 Jan 24. Biochemistry. 2025. PMID: 39854700 Free PMC article.
-
Generation of ribosomal protein S1 mutants for improving of expression of difficult to translate mRNAs.Appl Microbiol Biotechnol. 2025 Jan 23;109(1):20. doi: 10.1007/s00253-025-13406-4. Appl Microbiol Biotechnol. 2025. PMID: 39847144 Free PMC article.
-
Ribosomal mutations enable a switch between high fitness and high stress resistance in Listeria monocytogenes.Front Microbiol. 2024 Mar 28;15:1355268. doi: 10.3389/fmicb.2024.1355268. eCollection 2024. Front Microbiol. 2024. PMID: 38605704 Free PMC article.
References
-
- Yusupova G.Z., Yusupov M.M., Cate J.H., Noller H.F.. The path of messenger RNA through the ribosome. Cell. 2001; 106:233–241. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

