Red light-emitting short Mango-based system enables tracking a mycobacterial small noncoding RNA in infected macrophages
- PMID: 36840712
- PMCID: PMC10085697
- DOI: 10.1093/nar/gkad100
Red light-emitting short Mango-based system enables tracking a mycobacterial small noncoding RNA in infected macrophages
Abstract
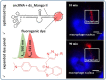
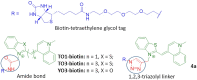
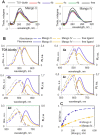
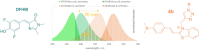
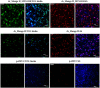
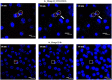
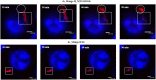
Progress in RNA metabolism and function studies relies largely on molecular imaging systems, including those comprising a fluorogenic dye and an aptamer-based fluorescence-activating tag. G4 aptamers of the Mango family, typically combined with a duplex/hairpin scaffold, activate the fluorescence of a green light-emitting dye TO1-biotin and hold great promise for intracellular RNA tracking. Here, we report a new Mango-based imaging platform. Its key advantages are the tunability of spectral properties and applicability for visualization of small RNA molecules that require minimal tag size. The former advantage is due to an expanded (green-to-red-emitting) palette of TO1-inspired fluorogenic dyes, and the truncated duplex scaffold ensures the latter. To illustrate the applicability of the improved platform, we tagged Mycobacterium tuberculosis sncRNA with the shortened aptamer-scaffold tag. Then, we visualized it in bacteria and bacteria-infected macrophages using the new red light-emitting Mango-activated dye.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Use of Genetically Encoded Fluorescent Aptamers for Visualization of Mycobacterium tuberculosis Small RNA MTS1338 in Infected Macrophages.Dokl Biochem Biophys. 2020 Jul;493(1):185-189. doi: 10.1134/S1607672920040055. Epub 2020 Sep 7. Dokl Biochem Biophys. 2020. PMID: 32894461
-
Double-stemmed and split structural variants of fluorescent RNA Mango aptamers.RNA. 2023 Sep;29(9):1355-1364. doi: 10.1261/rna.079651.123. Epub 2023 Jun 2. RNA. 2023. PMID: 37268327 Free PMC article.
-
Structure and functional reselection of the Mango-III fluorogenic RNA aptamer.Nat Chem Biol. 2019 May;15(5):472-479. doi: 10.1038/s41589-019-0267-9. Epub 2019 Apr 15. Nat Chem Biol. 2019. PMID: 30992561 Free PMC article.
-
In Vivo Production of RNA Aptamers and Nanoparticles: Problems and Prospects.Molecules. 2021 Mar 6;26(5):1422. doi: 10.3390/molecules26051422. Molecules. 2021. PMID: 33800717 Free PMC article. Review.
-
Light-Up RNA Aptamers and Their Cognate Fluorogens: From Their Development to Their Applications.Int J Mol Sci. 2017 Dec 23;19(1):44. doi: 10.3390/ijms19010044. Int J Mol Sci. 2017. PMID: 29295531 Free PMC article. Review.
Cited by
-
Structure-Informed Design of an Ultra Bright RNA-activated Fluorophore.Res Sq [Preprint]. 2024 Aug 5:rs.3.rs-4750449. doi: 10.21203/rs.3.rs-4750449/v1. Res Sq. 2024. Update in: Nat Chem. 2025 Aug;17(8):1188-1195. doi: 10.1038/s41557-025-01832-w. PMID: 39149476 Free PMC article. Updated. Preprint.
-
Structure-informed design of an ultrabright RNA-activated fluorophore.Nat Chem. 2025 Aug;17(8):1188-1195. doi: 10.1038/s41557-025-01832-w. Epub 2025 May 28. Nat Chem. 2025. PMID: 40437193 Free PMC article.
References
-
- Zhang Z., Zhang J., Diao L., Han L.. Small non-coding rnas in human cancer: function, clinical utility, and characterization. Oncogene. 2021; 40:1570–1577. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

