Actinobacillus pleuropneumoniae encodes multiple phase-variable DNA methyltransferases that control distinct phasevarions
- PMID: 36840716
- PMCID: PMC10123105
- DOI: 10.1093/nar/gkad091
Actinobacillus pleuropneumoniae encodes multiple phase-variable DNA methyltransferases that control distinct phasevarions
Abstract
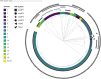
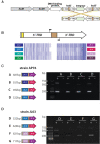
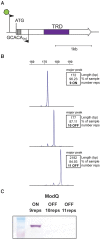
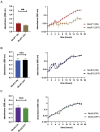
Actinobacillus pleuropneumoniae is the cause of porcine pleuropneumonia, a severe respiratory tract infection that is responsible for major economic losses to the swine industry. Many host-adapted bacterial pathogens encode systems known as phasevarions (phase-variable regulons). Phasevarions result from variable expression of cytoplasmic DNA methyltransferases. Variable expression results in genome-wide methylation differences within a bacterial population, leading to altered expression of multiple genes via epigenetic mechanisms. Our examination of a diverse population of A. pleuropneumoniae strains determined that Type I and Type III DNA methyltransferases with the hallmarks of phase variation were present in this species. We demonstrate that phase variation is occurring in these methyltransferases, and show associations between particular Type III methyltransferase alleles and serovar. Using Pacific BioSciences Single-Molecule, Real-Time (SMRT) sequencing and Oxford Nanopore sequencing, we demonstrate the presence of the first ever characterised phase-variable, cytosine-specific Type III DNA methyltransferase. Phase variation of distinct Type III DNA methyltransferase in A. pleuropneumoniae results in the regulation of distinct phasevarions, and in multiple phenotypic differences relevant to pathobiology. Our characterisation of these newly described phasevarions in A. pleuropneumoniae will aid in the selection of stably expressed antigens, and direct and inform development of a rationally designed subunit vaccine against this major veterinary pathogen.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Gottschalk M., Broes A.. Diseases of Swine. 2019; 749–766.
-
- Chiers K., Donné E., Van Overbeke I., Ducatelle R., Haesebrouck F.. Actinobacillus pleuropneumoniae infections in closed swine herds: infection patterns and serological profiles. Vet. Microbiol. 2002; 85:343–352. - PubMed
-
- Chiers K., Van Overbeke I., Donné E., Baele M., Ducatelle R., De Baere T., Haesebrouck F.. Detection of Actinobacillus pleuropneumoniae in cultures from nasal and tonsillar swabs of pigs by a PCR assay based on the nucleotide sequence of a dsbE-like gene. Vet. Microbiol. 2001; 83:147–159. - PubMed
-
- Vigre H., Angen Ø., Barfod K., Lavritsen D.T., Sørensen V.. Transmission of Actinobacillus pleuropneumoniae in pigs under field-like conditions: emphasis on tonsillar colonisation and passively acquired colostral antibodies. Vet. Microbiol. 2002; 89:151–159. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

