Computational methods and challenges in analyzing intratumoral microbiome data
- PMID: 36841736
- PMCID: PMC10272078
- DOI: 10.1016/j.tim.2023.01.011
Computational methods and challenges in analyzing intratumoral microbiome data
Abstract
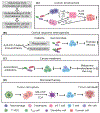
The human microbiome is intimately related to cancer biology and plays a vital role in the efficacy of cancer treatments, including immunotherapy. Extraordinary evidence has revealed that several microbes influence tumor development through interaction with the host immune system, that is, immuno-oncology-microbiome (IOM). This review focuses on the intratumoral microbiome in IOM and describes the available data and computational methods for discovering biological insights of microbial profiling from host bulk, single-cell, and spatial sequencing data. Critical challenges in data analysis and integration are discussed. Specifically, the microorganisms associated with cancer and cancer treatment in the context of IOM are collected and integrated from the literature. Lastly, we provide our perspectives for future directions in IOM research.
Keywords: cancer treatment and diagnosis; computational methods; immuno-oncology-microbiome; immunotherapy; intratumoral microbiome.
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests No interests are declared.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

