Systematic diet composition swap in a mouse genome-scale metabolic model reveals determinants of obesogenic diet metabolism in liver cancer
- PMID: 36844450
- PMCID: PMC9947310
- DOI: 10.1016/j.isci.2023.106040
Systematic diet composition swap in a mouse genome-scale metabolic model reveals determinants of obesogenic diet metabolism in liver cancer
Abstract
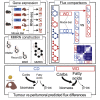
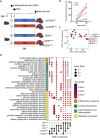
Dietary nutrient availability and gene expression, together, influence tissue metabolic activity. Here, we explore whether altering dietary nutrient composition in the context of mouse liver cancer suffices to overcome chronic gene expression changes that arise from tumorigenesis and western-style diet (WD). We construct a mouse genome-scale metabolic model and estimate metabolic fluxes in liver tumors and non-tumoral tissue after computationally varying the composition of input diet. This approach, called Systematic Diet Composition Swap (SyDiCoS), revealed that, compared to a control diet, WD increases production of glycerol and succinate irrespective of specific tissue gene expression patterns. Conversely, differences in fatty acid utilization pathways between tumor and non-tumor liver are amplified with WD by both dietary carbohydrates and lipids together. Our data suggest that combined dietary component modifications may be required to normalize the distinctive metabolic patterns that underlie selective targeting of tumor metabolism.
Keywords: Biological sciences; Cancer; Cellular physiology; Physiology.
© 2023 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Burton A., Tataru D., Driver R.J., Bird T.G., Huws D., Wallace D., Cross T.J.S., Rowe I.A., Alexander G., Marshall A., HCC-UK/BASL/NCRAS Partnership Steering Group Primary liver cancer in the UK: incidence, incidence-based mortality, and survival by subtype, sex, and nation. JHEP Rep. 2021;3:100232. doi: 10.1016/j.jhepr.2021.100232. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

