In Silico Discovery of Small Molecule Modulators Targeting the Achilles' Heel of SARS-CoV-2 Spike Protein
- PMID: 36844485
- PMCID: PMC9924089
- DOI: 10.1021/acscentsci.2c01190
In Silico Discovery of Small Molecule Modulators Targeting the Achilles' Heel of SARS-CoV-2 Spike Protein
Abstract
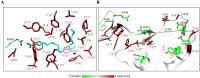
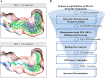
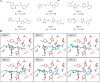
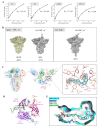
The spike protein of SARS-CoV-2 has been a promising target for developing vaccines and therapeutics due to its crucial role in the viral entry process. Previously reported cryogenic electron microscopy (cryo-EM) structures have revealed that free fatty acids (FFA) bind with SARS-CoV-2 spike protein, stabilizing its closed conformation and reducing its interaction with the host cell target in vitro. Inspired by these, we utilized a structure-based virtual screening approach against the conserved FFA-binding pocket to identify small molecule modulators of SARS-CoV-2 spike protein, which helped us identify six hits with micromolar binding affinities. Further evaluation of their commercially available and synthesized analogs enabled us to discover a series of compounds with better binding affinities and solubilities. Notably, our identified compounds exhibited similar binding affinities against the spike proteins of the prototypic SARS-CoV-2 and a currently circulating Omicron BA.4 variant. Furthermore, the cryo-EM structure of the compound SPC-14 bound spike revealed that SPC-14 could shift the conformational equilibrium of the spike protein toward the closed conformation, which is human ACE2 (hACE2) inaccessible. Our identified small molecule modulators targeting the conserved FFA-binding pocket could serve as the starting point for the future development of broad-spectrum COVID-19 intervention treatments.
© 2023 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Similar articles
-
A multiple-step in silico screening protocol to identify allosteric inhibitors of Spike-hACE2 binding.Phys Chem Chem Phys. 2022 Feb 16;24(7):4305-4316. doi: 10.1039/d1cp04736a. Phys Chem Chem Phys. 2022. PMID: 35107459
-
VE607 stabilizes SARS-CoV-2 Spike in the "RBD-up" conformation and inhibits viral entry.iScience. 2022 Jul 15;25(7):104528. doi: 10.1016/j.isci.2022.104528. Epub 2022 Jun 3. iScience. 2022. PMID: 35677392 Free PMC article.
-
In Silico Screening of Bioactive Compounds of Representative Seaweeds to Inhibit SARS-CoV-2 ACE2-Bound Omicron B.1.1.529 Spike Protein Trimer.Mar Drugs. 2022 Feb 17;20(2):148. doi: 10.3390/md20020148. Mar Drugs. 2022. PMID: 35200677 Free PMC article.
-
COVID-19 Coronavirus spike protein analysis for synthetic vaccines, a peptidomimetic antagonist, and therapeutic drugs, and analysis of a proposed achilles' heel conserved region to minimize probability of escape mutations and drug resistance.Comput Biol Med. 2020 Jun;121:103749. doi: 10.1016/j.compbiomed.2020.103749. Epub 2020 Apr 11. Comput Biol Med. 2020. PMID: 32568687 Free PMC article. Review.
-
The expression of hACE2 receptor protein and its involvement in SARS-CoV-2 entry, pathogenesis, and its application as potential therapeutic target.Tumour Biol. 2021;43(1):177-196. doi: 10.3233/TUB-200084. Tumour Biol. 2021. PMID: 34420993 Review.
Cited by
-
Different In Silico Approaches Using Heterocyclic Derivatives against the Binding between Different Lineages of SARS-CoV-2 and ACE2.Molecules. 2023 May 5;28(9):3908. doi: 10.3390/molecules28093908. Molecules. 2023. PMID: 37175318 Free PMC article.
-
Structure-based discovery of forsythoside A as an effective inhibitor to reduce the neurotoxicity of amyloid β.Commun Chem. 2025 Jul 4;8(1):196. doi: 10.1038/s42004-025-01587-y. Commun Chem. 2025. PMID: 40615672 Free PMC article.
-
Identification of Potential Lead Compounds Targeting Novel Druggable Cavity of SARS-CoV-2 Spike Trimer by Molecular Dynamics Simulations.Int J Mol Sci. 2023 Mar 27;24(7):6281. doi: 10.3390/ijms24076281. Int J Mol Sci. 2023. PMID: 37047254 Free PMC article.
-
Exploring Binding Pockets in the Conformational States of the SARS-CoV-2 Spike Trimers for the Screening of Allosteric Inhibitors Using Molecular Simulations and Ensemble-Based Ligand Docking.Int J Mol Sci. 2024 May 1;25(9):4955. doi: 10.3390/ijms25094955. Int J Mol Sci. 2024. PMID: 38732174 Free PMC article.
-
Spike Protein Mutation-Induced Changes in the Kinetic and Thermodynamic Behavior of Its Receptor Binding Domains Explain Their Higher Propensity to Attain Open States in SARS-CoV-2 Variants of Concern.ACS Cent Sci. 2023 Sep 21;9(10):1894-1904. doi: 10.1021/acscentsci.3c00810. eCollection 2023 Oct 25. ACS Cent Sci. 2023. PMID: 37901170 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

