Comprehensive Structure-Activity Relationship Studies of Cepafungin Enabled by Biocatalytic C-H Oxidations
- PMID: 36844499
- PMCID: PMC9951290
- DOI: 10.1021/acscentsci.2c01219
Comprehensive Structure-Activity Relationship Studies of Cepafungin Enabled by Biocatalytic C-H Oxidations
Abstract
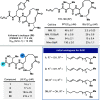
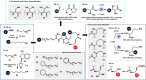
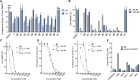
The cepafungins are a class of highly potent and selective eukaryotic proteasome inhibitor natural products with potential to treat refractory multiple myeloma and other cancers. The structure-activity relationship of the cepafungins is not fully understood. This Article chronicles the development of a chemoenzymatic approach to cepafungin I. A failed initial route involving derivatization of pipecolic acid prompted us to examine the biosynthetic pathway for the production of 4-hydroxylysine, which culminated in the development of a 9-step synthesis of cepafungin I. An alkyne-tagged analogue enabled chemoproteomic studies of cepafungin and comparison of its effects on global protein expression in human multiple myeloma cells to the clinical drug bortezomib. A preliminary series of analogues elucidated critical determinants of potency in proteasome inhibition. Herein we report the chemoenzymatic syntheses of 13 additional analogues of cepafungin I guided by a proteasome-bound crystal structure, 5 of which are more potent than the natural product. The lead analogue was found to have 7-fold greater proteasome β5 subunit inhibitory activity and has been evaluated against several multiple myeloma and mantle cell lymphoma cell lines in comparison to the clinical drug bortezomib.
© 2023 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare the following competing financial interest(s): A. Amatuni, A. Shuster, A. Adibekian, and H. Renata have applied for a provisional patent for this work.
Figures





Similar articles
-
Concise Chemoenzymatic Total Synthesis and Identification of Cellular Targets of Cepafungin I.Cell Chem Biol. 2020 Oct 15;27(10):1318-1326.e18. doi: 10.1016/j.chembiol.2020.07.012. Epub 2020 Aug 6. Cell Chem Biol. 2020. PMID: 32763140 Free PMC article.
-
Syrbactin proteasome inhibitor TIR-199 overcomes bortezomib chemoresistance and inhibits multiple myeloma tumor growth in vivo.Leuk Res. 2020 Jan;88:106271. doi: 10.1016/j.leukres.2019.106271. Epub 2019 Nov 12. Leuk Res. 2020. PMID: 31778912 Free PMC article.
-
The emerging role of bortezomib in the treatment of indolent non-Hodgkin's and mantle cell lymphomas.Curr Treat Options Oncol. 2004 Aug;5(4):269-81. doi: 10.1007/s11864-004-0018-2. Curr Treat Options Oncol. 2004. PMID: 15233904 Review.
-
Optical Control of Proteasomal Protein Degradation with a Photoswitchable Lipopeptide.Angew Chem Int Ed Engl. 2024 Feb 19;63(8):e202314791. doi: 10.1002/anie.202314791. Epub 2024 Jan 16. Angew Chem Int Ed Engl. 2024. PMID: 38109686 Free PMC article.
-
Proteasome inhibitors in the treatment of B-cell malignancies.Clin Lymphoma. 2002 Jun;3(1):49-55. doi: 10.3816/clm.2002.n.011. Clin Lymphoma. 2002. PMID: 12141956 Review.
Cited by
-
Multiplexed Assessment of Promiscuous Non-Canonical Amino Acid Synthase Activity in a Pyridoxal Phosphate-Dependent Protein Family.ACS Catal. 2023 Sep 1;13(17):11644-11655. doi: 10.1021/acscatal.3c02498. Epub 2023 Aug 21. ACS Catal. 2023. PMID: 37720819 Free PMC article.
-
Non-Native Site-Selective Enzyme Catalysis.Chem Rev. 2023 Aug 23;123(16):10381-10431. doi: 10.1021/acs.chemrev.3c00215. Epub 2023 Jul 31. Chem Rev. 2023. PMID: 37524057 Free PMC article. Review.
-
Biocatalytic C-H oxidation meets radical cross-coupling: Simplifying complex piperidine synthesis.Science. 2024 Dec 20;386(6728):1421-1427. doi: 10.1126/science.adr9368. Epub 2024 Dec 19. Science. 2024. PMID: 39700271
-
Advances, opportunities, and challenges in methods for interrogating the structure activity relationships of natural products.Nat Prod Rep. 2024 Oct 17;41(10):1543-1578. doi: 10.1039/d4np00009a. Nat Prod Rep. 2024. PMID: 38912779 Free PMC article. Review.
-
Computational analysis of antimicrobial peptides targeting key receptors in infection-related cardiovascular diseases: molecular docking and dynamics insights.Sci Rep. 2025 Mar 14;15(1):8896. doi: 10.1038/s41598-025-93683-1. Sci Rep. 2025. PMID: 40087360 Free PMC article.
References
-
- A Phase III Trial of With Marizomib in Patients With Newly Diagnosed Glioblastoma (MIRAGE); ClinicalTrials.gov identifier NCT03345095. https://clinicaltrials.gov/ct2/show/NCT03345095 (first posted November 17, 2017; last update posted October 6, 2020).
-
- Weyburne E. S.; Wilkins O. M.; Sha Z.; Williams D. A.; Pletnev A. A.; de Bruin G.; Overkleeft H. S.; Goldberg A. L.; Cole M. D.; Kisselev A. F. Inhibition of the Proteasome Β2 Site Sensitizes Triple-Negative Breast Cancer Cells to Β5 Inhibitors and Suppresses Nrf1 Activation. Cell Chem. Biol. 2017, 24 (2), 218–230. 10.1016/j.chembiol.2016.12.016. - DOI - PMC - PubMed
- Oerlemans R.; Franke N. E.; Assaraf Y. G.; Cloos J.; Van Zantwijk I.; Berkers C. R.; Scheffer G. L.; Debipersad K.; Vojtekova K.; Lemos C.; et al. Molecular Basis of Bortezomib Resistance: Proteasome Subunit 2 5 (PSMB5) Gene Mutation and Overexpression of PSMB5 Protein. Blood 2008, 112 (6), 2489–2499. 10.1182/blood-2007-08-104950. - DOI - PubMed
- Besse A.; Besse L.; Kraus M.; Mendez-Lopez M.; Bader J.; Xin B. T.; de Bruin G.; Maurits E.; Overkleeft H. S.; Driessen C. Proteasome Inhibition in Multiple Myeloma: Head-to-Head Comparison of Currently Available Proteasome Inhibitors. Cell Chem. Biol. 2019, 26 (3), 340–351. 10.1016/j.chembiol.2018.11.007. - DOI - PubMed
-
- Clerc J.; Groll M.; Illich D. J.; Bachmann A. S.; Huber R.; Schellenberg B.; Dudler R.; Kaiser M. Synthetic and Structural Studies on Syringolin A and B Reveal Critical Determinants of Selectivity and Potency of Proteasome Inhibition. Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (16), 6507–6512. 10.1073/pnas.0901982106. - DOI - PMC - PubMed
- Clerc J.; Schellenberg B.; Groll M.; Bachmann A. S.; Huber R.; Dudler R.; Kaiser M. Convergent Synthesis and Biological Evaluation of Syringolin A and Derivatives as Eukaryotic 20S Proteasome Inhibitors. Eur. J. Org. Chem. 2010, 2010 (21), 3991–4003. 10.1002/ejoc.201000317. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

