Data-Independent Acquisition Mass Spectrometry of EPS-Urine Coupled to Machine Learning: A Predictive Model for Prostate Cancer
- PMID: 36844540
- PMCID: PMC9948177
- DOI: 10.1021/acsomega.2c05487
Data-Independent Acquisition Mass Spectrometry of EPS-Urine Coupled to Machine Learning: A Predictive Model for Prostate Cancer
Abstract
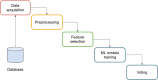
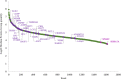
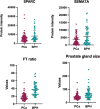
Prostate cancer (PCa) is annually the most frequently diagnosed cancer in the male population. To date, the diagnostic path for PCa detection includes the dosage of serum prostate-specific antigen (PSA) and the digital rectal exam (DRE). However, PSA-based screening has insufficient specificity and sensitivity; besides, it cannot discriminate between the aggressive and indolent types of PCa. For this reason, the improvement of new clinical approaches and the discovery of new biomarkers are necessary. In this work, expressed prostatic secretion (EPS)-urine samples from PCa patients and benign prostatic hyperplasia (BPH) patients were analyzed with the aim of detecting differentially expressed proteins between the two analyzed groups. To map the urinary proteome, EPS-urine samples were analyzed by data-independent acquisition (DIA), a high-sensitivity method particularly suitable for detecting proteins at low abundance. Overall, in our analysis, 2615 proteins were identified in 133 EPS-urine specimens obtaining the highest proteomic coverage for this type of sample; of these 2615 proteins, 1670 were consistently identified across the entire data set. The matrix containing the quantified proteins in each patient was integrated with clinical parameters such as the PSA level and gland size, and the complete matrix was analyzed by machine learning algorithms (by exploiting 90% of samples for training/testing using a 10-fold cross-validation approach, and 10% of samples for validation). The best predictive model was based on the following components: semaphorin-7A (sema7A), secreted protein acidic and rich in cysteine (SPARC), FT ratio, and prostate gland size. The classifier could predict disease conditions (BPH, PCa) correctly in 83% of samples in the validation set. Data are available via ProteomeXchange with the identifier PXD035942.
© 2023 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Similar articles
-
Development of a predictive model to distinguish prostate cancer from benign prostatic hyperplasia by integrating serum glycoproteomics and clinical variables.Clin Proteomics. 2023 Nov 21;20(1):52. doi: 10.1186/s12014-023-09439-4. Clin Proteomics. 2023. PMID: 37990292 Free PMC article.
-
Identification of prostate-enriched proteins by in-depth proteomic analyses of expressed prostatic secretions in urine.J Proteome Res. 2012 Apr 6;11(4):2386-96. doi: 10.1021/pr2011236. Epub 2012 Feb 29. J Proteome Res. 2012. PMID: 22339264 Free PMC article.
-
More advantages in detecting bone and soft tissue metastases from prostate cancer using 18F-PSMA PET/CT.Hell J Nucl Med. 2019 Jan-Apr;22(1):6-9. doi: 10.1967/s002449910952. Epub 2019 Mar 7. Hell J Nucl Med. 2019. PMID: 30843003
-
Urinary microRNA-based signature improves accuracy of detection of clinically relevant prostate cancer within the prostate-specific antigen grey zone.Mol Med Rep. 2016 Jun;13(6):4549-60. doi: 10.3892/mmr.2016.5095. Epub 2016 Apr 8. Mol Med Rep. 2016. PMID: 27081843 Free PMC article.
-
Proteomics in diagnosis of prostate cancer.Pril (Makedon Akad Nauk Umet Odd Med Nauki). 2015;36(1):5-36. Pril (Makedon Akad Nauk Umet Odd Med Nauki). 2015. PMID: 26076772 Review.
Cited by
-
Development of a predictive model to distinguish prostate cancer from benign prostatic hyperplasia by integrating serum glycoproteomics and clinical variables.Clin Proteomics. 2023 Nov 21;20(1):52. doi: 10.1186/s12014-023-09439-4. Clin Proteomics. 2023. PMID: 37990292 Free PMC article.
-
Comparative efficacy of radical prostatectomy and radiotherapy in the treatment of high-risk prostate cancer.Technol Health Care. 2024;32(6):4671-4679. doi: 10.3233/THC-240910. Technol Health Care. 2024. PMID: 39093097 Free PMC article.
-
Recent progress in mass spectrometry-based urinary proteomics.Clin Proteomics. 2024 Feb 22;21(1):14. doi: 10.1186/s12014-024-09462-z. Clin Proteomics. 2024. PMID: 38389064 Free PMC article. Review.
-
Multiplexed quantitative proteomics in prostate cancer biomarker development.Adv Cancer Res. 2024;161:31-69. doi: 10.1016/bs.acr.2024.04.003. Epub 2024 Apr 25. Adv Cancer Res. 2024. PMID: 39032952 Free PMC article. Review.
-
Machine learning pipeline to analyze clinical and proteomics data: experiences on a prostate cancer case.BMC Med Inform Decis Mak. 2024 Apr 8;24(1):93. doi: 10.1186/s12911-024-02491-6. BMC Med Inform Decis Mak. 2024. PMID: 38584282 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

