(8-Hydroxyquinoline) Gallium(III) Complex with High Antineoplastic Efficacy for Treating Colon Cancer via Multiple Mechanisms
- PMID: 36844596
- PMCID: PMC9948165
- DOI: 10.1021/acsomega.2c07742
(8-Hydroxyquinoline) Gallium(III) Complex with High Antineoplastic Efficacy for Treating Colon Cancer via Multiple Mechanisms
Abstract
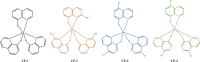
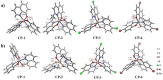
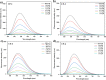
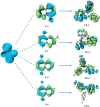
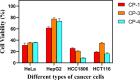
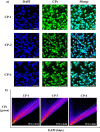
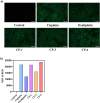
A series of (8-hydroxyquinoline) gallium(III) complexes (CP-1-4) was synthesized and characterized by single X-ray crystallography and density functional theory (DFT) calculation. The cytotoxicity of the four gallium complexes toward a human nonsmall cell lung cancer cell line (A549), human colon cancer cell line (HCT116), and human normal hepatocyte cell line (LO2) was evaluated using MTT assays. CP-4 exhibited excellent cytotoxicity against HCT116 cancer cells (IC50 = 1.2 ± 0.3 μM) and lower toxicity than cisplatin and oxaliplatin. We also evaluated the anticancer mechanism studies in cell uptake, reactive oxygen species analysis, cell cycle, wound-healing, and Western blotting assays. The results showed that CP-4 affected the expression of DNA-related proteins, which led to the apoptosis of cancer cells. Moreover, molecular docking tests of CP-4 were performed to predict other binding sites and to confirm its higher binding force to disulfide isomerase (PDI) proteins. The emissive properties of CP-4 suggest that this complex can be used for colon cancer diagnosis and treatment, as well as in vivo imaging. These results also provide a foundation for the development of gallium complexes as potent anticancer agents.
© 2023 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

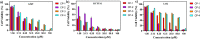




Similar articles
-
Novel and Versatile Imine-N-Heterocyclic Carbene Half-Sandwich Iridium(III) Complexes as Lysosome-Targeted Anticancer Agents.Inorg Chem. 2018 Sep 4;57(17):11087-11098. doi: 10.1021/acs.inorgchem.8b01656. Epub 2018 Aug 22. Inorg Chem. 2018. PMID: 30133276
-
Half-Lantern Cyclometalated Platinum(II) Complexes as Anticancer Agents: Molecular Docking, Apoptosis, Cell Cycle Analysis, and Cytotoxic Activity Evaluations.Anticancer Agents Med Chem. 2022;22(6):1149-1158. doi: 10.2174/1871520621666210713112105. Anticancer Agents Med Chem. 2022. PMID: 34259151
-
Gallium Metal-Organic Nanoparticles with Albumin-Stabilized and Loaded Graphene for Enhanced Delivery to HCT116 Cells.Int J Nanomedicine. 2023 Jan 13;18:225-241. doi: 10.2147/IJN.S386253. eCollection 2023. Int J Nanomedicine. 2023. PMID: 36660337 Free PMC article.
-
A significantly non-toxic novel Cobalt(III) Schiff base complex induces apoptosis via G2-M cell cycle arrest in human breast cancer cell line MCF-7.Life Sci. 2022 Nov 1;308:120963. doi: 10.1016/j.lfs.2022.120963. Epub 2022 Sep 13. Life Sci. 2022. PMID: 36113731
-
Thiosemicarbazone Cu(II) and Zn(II) complexes as potential anticancer agents: syntheses, crystal structure, DNA cleavage, cytotoxicity and apoptosis induction activity.J Inorg Biochem. 2014 Jul;136:13-23. doi: 10.1016/j.jinorgbio.2014.03.004. Epub 2014 Mar 16. J Inorg Biochem. 2014. PMID: 24690556
Cited by
-
Gallium complex K6 inhibits colorectal cancer by increasing ROS levels to induce DNA damage and enhance phosphatase and tensin homolog activity.MedComm (2020). 2024 Jul 24;5(8):e665. doi: 10.1002/mco2.665. eCollection 2024 Aug. MedComm (2020). 2024. PMID: 39049965 Free PMC article.
-
Highly Brominated Quinolines: Synthesis, Characterization, and Investigation of Anticancer Activities Supported by Molecular Dynamics.Chem Biol Drug Des. 2025 May;105(5):e70120. doi: 10.1111/cbdd.70120. Chem Biol Drug Des. 2025. PMID: 40329364 Free PMC article.
-
Responsive ZIF-90 nanocomposite material: targeted delivery of 10-hydroxycamptothecine to enhance the therapeutic effect of colon cancer (HCT116) cells.RSC Med Chem. 2024 May 9;15(8):2663-2676. doi: 10.1039/d3md00725a. eCollection 2024 Aug 14. RSC Med Chem. 2024. PMID: 39149092 Free PMC article.
-
First Gallium and Indium Crystal Structures of Curcuminoid Homoleptic Complexes: All-Different Ligand Stereochemistry and Cytotoxic Potential.Int J Mol Sci. 2023 Nov 15;24(22):16324. doi: 10.3390/ijms242216324. Int J Mol Sci. 2023. PMID: 38003515 Free PMC article.
References
-
- Karumban K. S.; Muley A.; Raut R.; Gupta P.; Giri B.; Kumbhakar S.; Misra A.; Maji S. Mononuclear Co(Ii) Polypyridyl Complexes: Synthesis, Molecular Structure, DNA Binding/Cleavage, Radical Scavenging, Docking Studies and Anticancer Activities. Dalt. Trans. 2022, 51, 7084–7099. 10.1039/D1DT04144D. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

