From rivers to ocean basins: The role of ocean barriers and philopatry in the genetic structuring of a cosmopolitan coastal predator
- PMID: 36844667
- PMCID: PMC9944188
- DOI: 10.1002/ece3.9837
From rivers to ocean basins: The role of ocean barriers and philopatry in the genetic structuring of a cosmopolitan coastal predator
Abstract
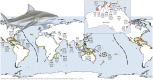
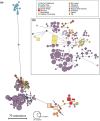
The Bull Shark (Carcharhinus leucas) faces varying levels of exploitation around the world due to its coastal distribution. Information regarding population connectivity is crucial to evaluate its conservation status and local fishing impacts. In this study, we sampled 922 putative Bull Sharks from 19 locations in the first global assessment of population structure of this cosmopolitan species. Using a recently developed DNA-capture approach (DArTcap), samples were genotyped for 3400 nuclear markers. Additionally, full mitochondrial genomes of 384 Indo-Pacific samples were sequenced. Reproductive isolation was found between and across ocean basins (eastern Pacific, western Atlantic, eastern Atlantic, Indo-West Pacific) with distinct island populations in Japan and Fiji. Bull Sharks appear to maintain gene flow using shallow coastal waters as dispersal corridors, whereas large oceanic distances and historical land-bridges act as barriers. Females tend to return to the same area for reproduction, making them more susceptible to local threats and an important focus for management actions. Given these behaviors, the exploitation of Bull Sharks from insular populations, such as Japan and Fiji, may instigate local decline that cannot readily be replenished by immigration, which can in turn affect ecosystem dynamics and functions. These data also supported the development of a genetic panel to ascertain the population of origin, which will be useful in monitoring the trade of fisheries products and assessing population-level impacts of this harvest.
Keywords: DArTseq; DNA forensics; close‐kin; genetic connectivity; mitogenome; provenance.
© 2023 The Authors. Ecology and Evolution published by John Wiley & Sons Ltd.
Conflict of interest statement
We note no conflict or competing interests among the authors in relation to the information provided in this manuscript.
Figures


References
-
- Anderson, E. C. , Waples, R. S. , & Kalinowski, S. T. (2008). An improved method for predicting the accuracy of genetic stock identification. Canadian Journal of Fisheries and Aquatic Sciences, 65(7), 1475–1486. 10.1139/F08-049 - DOI
-
- Avise, J. C. , Bowen, B. W. , Lamb, T. , Meylan, A. B. , & Bermingham, E. (1992). Mitochondrial DNA evolution at a turtle's pace: Evidence for low genetic variability and reduced microevolutionary rate in the Testudines. Molecular Biology and Evolution, 9(3), 457–473. 10.1093/oxfordjournals.molbev.a040735 - DOI - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

