Nectary development in Cleome violacea
- PMID: 36844906
- PMCID: PMC9949531
- DOI: 10.3389/fpls.2022.1085900
Nectary development in Cleome violacea
Abstract
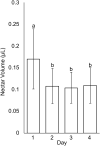
Nectaries are a promising frontier for plant evo-devo research, and are particularly fascinating given their diversity in form, position, and secretion methods across angiosperms. Emerging model systems permit investigations of the molecular basis for nectary development and nectar secretion across a range of taxa, which addresses fundamental questions about underlying parallelisms and convergence. Herein, we explore nectary development and nectar secretion in the emerging model taxa, Cleome violacea (Cleomaceae), which exhibits a prominent adaxial nectary. First, we characterized nectary anatomy and quantified nectar secretion to establish a foundation for quantitative and functional gene experiments. Next, we leveraged RNA-seq to establish gene expression profiles of nectaries across three key stages of development: pre-anthesis, anthesis, and post-fertilization. We then performed functional studies on five genes that were putatively involved in nectary and nectar formation: CvCRABSCLAW (CvCRC), CvAGAMOUS (CvAG), CvSHATTERPROOF (CvSHP), CvSWEET9, and a highly expressed but uncharacterized transcript. These experiments revealed a high degree of functional convergence to homologues from other core Eudicots, especially Arabidopsis. CvCRC, redundantly with CvAG and CvSHP, are required for nectary initiation. Concordantly, CvSWEET9 is essential for nectar formation and secretion, which indicates that the process is eccrine based in C. violacea. While demonstration of conservation is informative to our understanding of nectary evolution, questions remain. For example, it is unknown which genes are downstream of the developmental initiators CvCRC, CvAG, and CvSHP, or what role the TCP gene family plays in nectary initiation in this family. Further to this, we have initiated a characterization of associations between nectaries, yeast, and bacteria, but more research is required beyond establishing their presence. Cleome violacea is an excellent model for continued research into nectary development because of its conspicuous nectaries, short generation time, and close taxonomic distance to Arabidopsis.
Keywords: Cleomaceae; RNA-seq; VIGS; nectar; nectaries; parallel evolution; transcriptomics.
Copyright © 2023 Carey, Zenchyzen, Deneka and Hall.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures










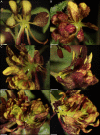
Similar articles
-
Comparative Nectary Morphology across Cleomaceae (Brassicales).Plants (Basel). 2023 Mar 10;12(6):1263. doi: 10.3390/plants12061263. Plants (Basel). 2023. PMID: 36986951 Free PMC article.
-
The developmental basis of floral nectary diversity and evolution.New Phytol. 2025 Jun;246(6):2462-2477. doi: 10.1111/nph.70141. Epub 2025 May 1. New Phytol. 2025. PMID: 40313027 Free PMC article. Review.
-
Transcriptomic and microstructural analyses in Liriodendron tulipifera Linn. reveal candidate genes involved in nectary development and nectar secretion.BMC Plant Biol. 2019 Dec 2;19(1):531. doi: 10.1186/s12870-019-2140-0. BMC Plant Biol. 2019. PMID: 31791230 Free PMC article.
-
Uncovering the Arabidopsis thaliana nectary transcriptome: investigation of differential gene expression in floral nectariferous tissues.BMC Plant Biol. 2009 Jul 15;9:92. doi: 10.1186/1471-2229-9-92. BMC Plant Biol. 2009. PMID: 19604393 Free PMC article.
-
Arabidopsis thaliana as a model for functional nectary analysis.Sex Plant Reprod. 2009 Dec;22(4):235-46. doi: 10.1007/s00497-009-0112-5. Epub 2009 Sep 1. Sex Plant Reprod. 2009. PMID: 20033445 Review.
Cited by
-
Shining a light on UV-fluorescent floral nectar after 50 years.Sci Rep. 2024 May 25;14(1):11992. doi: 10.1038/s41598-024-62626-7. Sci Rep. 2024. PMID: 38796543 Free PMC article.
-
CRABS CLAW-independent floral nectary development in Penstemon barbatus.Am J Bot. 2025 Jun;112(6):e70058. doi: 10.1002/ajb2.70058. Epub 2025 Jun 16. Am J Bot. 2025. PMID: 40524523 Free PMC article.
-
Comparative Nectary Morphology across Cleomaceae (Brassicales).Plants (Basel). 2023 Mar 10;12(6):1263. doi: 10.3390/plants12061263. Plants (Basel). 2023. PMID: 36986951 Free PMC article.
-
Can nectary structure in Laeliinae promote or constrain nectar secretion?BMC Plant Biol. 2025 Jun 7;25(1):772. doi: 10.1186/s12870-025-06810-5. BMC Plant Biol. 2025. PMID: 40483404 Free PMC article.
-
The developmental basis of floral nectary diversity and evolution.New Phytol. 2025 Jun;246(6):2462-2477. doi: 10.1111/nph.70141. Epub 2025 May 1. New Phytol. 2025. PMID: 40313027 Free PMC article. Review.
References
-
- Alvarez J., Smyth D. R. (1998). Genetic pathways controlling carpel development in Arabidopsis thaliana . J. Plant Res. 111, 295–298. doi: 10.1007/BF02512187 - DOI
-
- Alvarez J., Smyth D. R. (2002). CRABS CLAW and SPATULA genes regulate growth and pattern formation during gynoecium development in Arabidopsis thaliana . Int. J. Plant Sci. 163, 17–41. doi: 10.1086/324178 - DOI
LinkOut - more resources
Full Text Sources

