Evolution of increased positive charge on the SARS-CoV-2 spike protein may be adaptation to human transmission
- PMID: 36845032
- PMCID: PMC9937996
- DOI: 10.1016/j.isci.2023.106230
Evolution of increased positive charge on the SARS-CoV-2 spike protein may be adaptation to human transmission
Abstract
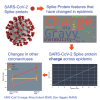
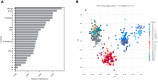
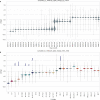
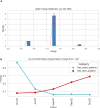
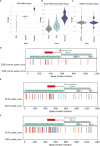
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to evolve and infect individuals. The exterior surface of the SARS-CoV-2 virion is dominated by the spike protein, and the current work examined spike protein biochemical features that have changed during the 3 years in which SARS-CoV-2 has infected humans. Our analysis identified a striking change in spike protein charge, from -8.3 in the original Lineage A and B viruses to -1.26 in most of the current Omicron viruses. We conclude that in addition to immune selection pressure, the evolution of SARS-CoV-2 has also altered viral spike protein biochemical properties, which may influence virion survival and promote transmission. Future vaccine and therapeutic development should also exploit and target these biochemical properties.
Keywords: Evolutionary biology; Virology.
© 2023 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29:463–476.e6. doi: 10.1016/j.chom.2021.02.003. - DOI - PMC - PubMed
-
- Greaney A.J., Starr T.N., Barnes C.O., Weisblum Y., Schmidt F., Caskey M., Gaebler C., Cho A., Agudelo M., Finkin S., et al. Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat. Commun. 2021;12:4196. doi: 10.1038/s41467-021-24435-8. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

