Osthole regulates N6-methyladenosine-modified TGM2 to inhibit the progression of rheumatoid arthritis and associated interstitial lung disease
- PMID: 36845072
- PMCID: PMC9945862
- DOI: 10.1002/mco2.219
Osthole regulates N6-methyladenosine-modified TGM2 to inhibit the progression of rheumatoid arthritis and associated interstitial lung disease
Abstract
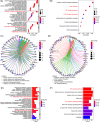
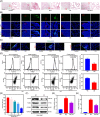
Rheumatoid arthritis (RA) is an inflammatory autoimmune disease, and RA interstitial lung disease (ILD) is a severe complication of RA. This investigation aims to determine the effect and underlying mechanism of osthole (OS), which could be extracted from Cnidium, Angelica, and Citrus plants and evaluate the role of transglutaminase 2 (TGM2) in RA and RA-ILD. In this work, OS downregulated TGM2 to exert its additive effect with methotrexate and suppress the proliferation, migration, and invasion of RA-fibroblast-like synoviocytes (FLS) by attenuating NF-κB signaling, resulting in the suppression of RA progression. Interestingly, WTAP-mediated N6-methyladenosine modification of TGM2 and Myc-mediated WTAP transcription cooperatively contributed to the formation of a TGM2/Myc/WTAP-positive feedback loop through upregulating NF-κB signaling. Moreover, OS could downregulate the activation of the TGM2/Myc/WTAP-positive feedback circuit. Furthermore, OS restrained the proliferation and polarization of M2 macrophages to inhibit the aggregation of lung interstitial CD11b+ macrophages, and the effectiveness and non-toxicity of OS in suppressing RA and RA-ILD progression were verified in vivo. Finally, bioinformatics analyses validated the importance and the clinical significance of the OS-regulated molecular network. Taken together, our work emphasized OS as an effective drug candidate and TGM2 as a promising target for RA and RA-ILD treatment.
Keywords: TGM2; interstitial lung disease; osthole; rheumatoid arthritis.
© 2023 The Authors. MedComm published by Sichuan International Medical Exchange & Promotion Association (SCIMEA) and John Wiley & Sons Australia, Ltd.
Conflict of interest statement
The authors declare no competing interest.
Figures






References
-
- Daraghmeh DN, King C, Wiese MD. A review of liquid biopsy as a tool to assess epigenetic, cfDNA and miRNA variability as methotrexate response predictors in patients with rheumatoid arthritis. Pharmacol Res. 2021;173:105887. - PubMed
-
- Wang W, Zhou H, Liu L. Side effects of methotrexate therapy for rheumatoid arthritis: a systematic review. Eur J Med Chem. 2018;158:502‐516. - PubMed
-
- Lin X, Tao C, Zhang R, Zhang M, Wang Q, Chen J. N6‐methyladenosine modification of TGM2 mRNA contributes to the inhibitory activity of sarsasapogenin in rheumatoid arthritis fibroblast‐like synoviocytes. Phytomedicine. 2022;95:153871. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
