High throughput sRNA sequencing revealed gene regulatory role mediated by pathogen-derived small RNAs during Sri Lankan Cassava Mosaic Virus infection in Cassava
- PMID: 36845076
- PMCID: PMC9950310
- DOI: 10.1007/s13205-023-03494-2
High throughput sRNA sequencing revealed gene regulatory role mediated by pathogen-derived small RNAs during Sri Lankan Cassava Mosaic Virus infection in Cassava
Abstract
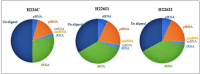
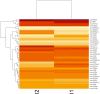
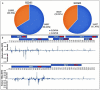
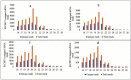
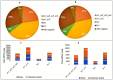
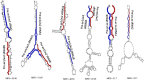
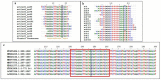
Small RNA (sRNA) mediated gene regulation during Sri Lankan Cassava Mosaic Virus (SLCMV) infection was studied from the Indian Cassava Cultivar H226. Our study generated high throughput sRNA dataset of 23.64 million reads from the control and SLCMV infected H226 leaf libraries. mes-miR9386 was detected as the most prominent miRNA expressed in control and infected leaf. Among the differentially expressed miRNAs, mes-miR156, mes- miR395 and mes-miR535a/b showed significant down regulation in the infected leaf. Genome-wide analysis of the three small RNA profiles revealed critical role of virus-derived small RNAs (vsRNAs) from the infected leaf tissues of H226. The vsRNAs were mapped to the bipartite SLCMV genome and high expression of siRNAs generated from the virus genomic region encoding AV1/AV2 genes in the infected leaf pointed towards the susceptibility of H226 cultivars to SLCMV. Furthermore, the sRNA reads mapped to the antisense strand of the SLCMV ORFs was higher than the sense strand. These vsRNAs were potential to target key host genes involved in virus interaction such as aldehyde dehydrogenase, ADP-ribosylation factor1 and ARF1-like GTP-binding proteins. The sRNAome-assisted analysis also revealed the origin of virus-encoded miRNAs from the SLCMV genome in the infected leaf. These virus-derived miRNAs were predicted to have hair-pin like secondary structures, and have different isoforms. Moreover, our study revealed that the pathogen sRNAs play a critical role in the infection process in H226 plants.
Supplementary information: The online version contains supplementary material available at 10.1007/s13205-023-03494-2.
Keywords: Cassava; H226; Sri Lankan Cassava Mosaic virus; Virus-derived small RNAs; miRNAs.
© King Abdulaziz City for Science and Technology 2023, Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Conflict of interest statement
Conflict of interestOn behalf of all authors, the corresponding author states that there is no conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

