Age-dependent Lamin changes induce cardiac dysfunction via dysregulation of cardiac transcriptional programs
- PMID: 36845078
- PMCID: PMC9956937
- DOI: 10.1038/s43587-022-00323-8
Age-dependent Lamin changes induce cardiac dysfunction via dysregulation of cardiac transcriptional programs
Abstract
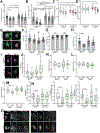
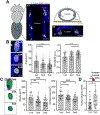
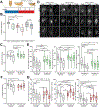
As we age, structural changes contribute to progressive decline in organ function, which in the heart act through poorly characterized mechanisms. Taking advantage of the short lifespan and conserved cardiac proteome of the fruit fly, we found that cardiomyocytes exhibit progressive loss of Lamin C (mammalian Lamin A/C homologue) with age, coincident with decreasing nuclear size and increasing nuclear stiffness. Premature genetic reduction of Lamin C phenocopies aging's effects on the nucleus, and subsequently decreases heart contractility and sarcomere organization. Surprisingly, Lamin C reduction downregulates myogenic transcription factors and cytoskeletal regulators, possibly via reduced chromatin accessibility. Subsequently, we find a role for cardiac transcription factors in regulating adult heart contractility and show that maintenance of Lamin C, and cardiac transcription factor expression, prevents age-dependent cardiac decline. Our findings are conserved in aged non-human primates and mice, demonstrating that age-dependent nuclear remodeling is a major mechanism contributing to cardiac dysfunction.
Figures
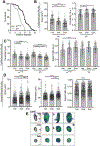
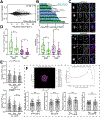

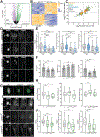
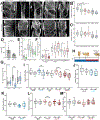






Comment in
-
Nuclear remodelling contributes to cardiac dysfunction during ageing.Nat Rev Cardiol. 2023 Mar;20(3):140. doi: 10.1038/s41569-023-00832-y. Nat Rev Cardiol. 2023. PMID: 36653464 No abstract available.
References
-
- Gilbert HTJ & Swift J The consequences of ageing, progeroid syndromes and cellular senescence on mechanotransduction and the nucleus. Exp Cell Res 378, 98–103 (2019). - PubMed
-
- CDC, N. Underlying Cause of Death 1999-2013 on CDC WONDER Online Database, released 2015. Data are from the Multiple Cause of Death Files, 1999-2013, as compiled from data provided by the 57 vital statistics jurisdictions through the Vital Statistics Cooperative . (2015).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

