Detailed characterization of SARS-CoV-2-specific T and B cells after infection or heterologous vaccination
- PMID: 36845156
- PMCID: PMC9947839
- DOI: 10.3389/fimmu.2023.1123724
Detailed characterization of SARS-CoV-2-specific T and B cells after infection or heterologous vaccination
Abstract
The formation of a robust long-term antigen (Ag)-specific memory, both humoral and cell-mediated, is created following severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection or vaccination. Here, by using polychromatic flow cytometry and complex data analyses, we deeply investigated the magnitude, phenotype, and functionality of SARS-CoV-2-specific immune memory in two groups of healthy subjects after heterologous vaccination compared to a group of subjects who recovered from SARS-CoV-2 infection. We find that coronavirus disease 2019 (COVID-19) recovered patients show different long-term immunological profiles compared to those of donors who had been vaccinated with three doses. Vaccinated individuals display a skewed T helper (Th)1 Ag-specific T cell polarization and a higher percentage of Ag-specific and activated memory B cells expressing immunoglobulin (Ig)G compared to those of patients who recovered from severe COVID-19. Different polyfunctional properties characterize the two groups: recovered individuals show higher percentages of CD4+ T cells producing one or two cytokines simultaneously, while the vaccinated are distinguished by highly polyfunctional populations able to release four molecules, namely, CD107a, interferon (IFN)-γ, tumor necrosis factor (TNF), and interleukin (IL)-2. These data suggest that functional and phenotypic properties of SARS-CoV-2 adaptive immunity differ in recovered COVID-19 individuals and vaccinated ones.
Keywords: B cells; SARS-CoV-2; T cells; antigen-specific response; cytokine; polyfunctionality.
Copyright © 2023 Lo Tartaro, Paolini, Mattioli, Swatler, Neroni, Borella, Santacroce, Di Nella, Gozzi, Busani, Cuccorese, Trenti, Meschiari, Guaraldi, Girardis, Mussini, Piwocka, Gibellini, Cossarizza and De Biasi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer AC declared a past co-authorship with some of the authors to the handling editor.
Figures


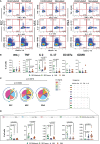

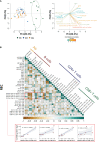
References
-
- Cohen KW, Linderman SL, Moodie Z, Czartoski J, Lai L, Mantus G, et al. . Longitudinal analysis shows durable and broad immune memory after SARS-CoV-2 infection with persisting antibody responses and memory b and T cells. Cell Rep Med (2021) 2(7):100354. doi: 10.1016/j.xcrm.2021.100354 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

