Autoantibody repertoire characterization provides insight into the pathogenesis of monogenic and polygenic autoimmune diseases
- PMID: 36845162
- PMCID: PMC9955420
- DOI: 10.3389/fimmu.2023.1106537
Autoantibody repertoire characterization provides insight into the pathogenesis of monogenic and polygenic autoimmune diseases
Abstract
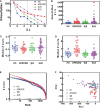
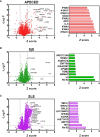
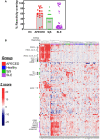
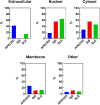
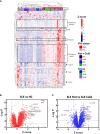
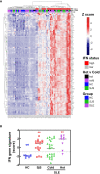
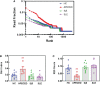
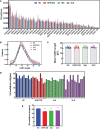
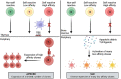
Autoimmune diseases vary in the magnitude and diversity of autoantibody profiles, and these differences may be a consequence of different types of breaks in tolerance. Here, we compared the disparate autoimmune diseases autoimmune polyendocrinopathy-candidiasis-ecto-dermal dystrophy (APECED), systemic lupus erythematosus (SLE), and Sjogren's syndrome (SjS) to gain insight into the etiology of breaks in tolerance triggering autoimmunity. APECED was chosen as a prototypical monogenic disease with organ-specific pathology while SjS and SLE represent polygenic autoimmunity with focal or systemic disease. Using protein microarrays for autoantibody profiling, we found that APECED patients develop a focused but highly reactive set of shared mostly anti-cytokine antibodies, while SLE patients develop broad and less expanded autoantibody repertoires against mostly intracellular autoantigens. SjS patients had few autoantibody specificities with the highest shared reactivities observed against Ro-52 and La. RNA-seq B-cell receptor analysis revealed that APECED samples have fewer, but highly expanded, clonotypes compared with SLE samples containing a diverse, but less clonally expanded, B-cell receptor repertoire. Based on these data, we propose a model whereby the presence of autoreactive T-cells in APECED allows T-dependent B-cell responses against autoantigens, while SLE is driven by breaks in peripheral B-cell tolerance and extrafollicular B-cell activation. These results highlight differences in the autoimmunity observed in several monogenic and polygenic disorders and may be generalizable to other autoimmune diseases.
Keywords: B cell receptor (BCR); Sjogren's syndrome; autoantibody; autoimmune disease (AD); autoimmune polyendocrinopathy candidiasis ecotodermal dystrophy (APECED); systemic lupus erythematosus (SLE).
Copyright © 2023 Clarke, Du, Kumar, Okitsu, Schuette, An, Zhang, Tzvetkov, Jensen, Niewold, Ferre, Nardone, Lionakis, Vlach, DeMartino and Bender.
Conflict of interest statement
TC, PD, SK, SO, MS, QA, JZ, ET, JV, JD, AB are/have been employees of EMD Serono, Billerica MA, USA or Merck KGaA, Darmstadt, Germany. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest. The authors declare that this study received funding from EMD Serono Inc and Merck KGaA. The funders were involved in the study design, collection, analysis, interpretation of data, the writing of this article and the decision to submit it for publication.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

